Back to Journals » OncoTargets and Therapy » Volume 7
Efficient utilization of EBUS-TBNA samples for both diagnosis and molecular analyses
Authors Oezkan F, Khan AM, Zarogoulidis P, Hohenforst-Schmidt W, Theegarten D, Yasufuku K, Nakajima T, Freitag L, Darwiche K
Received 20 August 2014
Accepted for publication 1 October 2014
Published 10 November 2014 Volume 2014:7 Pages 2061—2065
DOI https://doi.org/10.2147/OTT.S72974
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Faris Farassati
F Oezkan,1 AM Khan,2 P Zarogoulidis,3 W Hohenforst-Schmidt,4 D Theegarten,5 K Yasufuku,2 T Nakajima,6 L Freitag,1 K Darwiche1
1Department of Interventional Pneumology, Ruhrlandklinik, University Hospital Essen, University of Duisburg-Essen, Essen, Germany; 2Division of Thoracic Surgery, Toronto General Hospital, University Health Network, University of Toronto, Toronto, ON, Canada; 3Pulmonary Department-Oncology Unit, 'G Papanikolaou' General Hospital, Aristotle University of Thessaloniki, Thessaloniki, Greece; 4II Medical Clinic, Coburg Hospital, University of Wuerzburg, Coburg, Germany; 5Institute of Pathology, University Hospital of Essen, University of Duisburg-Essen, Essen, Germany; 6Department of General Thoracic Surgery, Graduate School of Medicine, Chiba University, Chiba, Japan
Background: The assessment of an increasing number of molecular markers is becoming a standard requirement from endobronchial ultrasound-guided transbronchial needle aspiration (EBUS-TBNA) specimens. However, it is unclear how many needle passes should be performed and the amount of lung cancer cells that should be sent for molecular analyses. The objective of this study was to determine if it is feasible to divide the material obtained by EBUS-TBNA to allow for molecular analysis without compromising the accuracy of mediastinal staging.
Objective: We aimed to determine if dividing EBUS-TBNA specimens has a negative impact on either histopathological diagnosis or molecular analysis.
Methods: EBUS-TBNA was performed in 249 enlarged lymph nodes. Negative or ambiguous histopathological results were confirmed by surgical means and clinical follow-up over 6 months. The tissue obtained by EBUS-TBNA was placed onto a glass slide and divided for histopathological workup and molecular analysis. The number of passes was recorded. Both the accuracy of the mediastinal lymph node staging and the applicability of the sample division for molecular analysis were assessed.
Results: Each lymph node was punctured an average of 3.18 times and division of the obtained material for diagnosis and molecular analysis was feasible in all cases. The sensitivity and accuracy of the mediastinal lymph node staging were 96.6% and 97.6%, respectively. A cytokeratin (CK)-19-mRNA concentration–based molecular test was feasible in 74.1% of cases.
Conclusion: Dividing EBUS-TBNA samples for both histopathological diagnosis and molecular testing is feasible and does not compromise the accuracy of mediastinal staging. This method may be an alternative to taking additional needle passes for molecular analyses.
Keywords: lung cancer, molecular marker, CK-19-mRNA, lymph node sampling
Introduction
Accurate mediastinal lymph node staging is an essential preoperative component in a lung cancer workup. Endobronchial ultrasound-guided transbronchial needle aspiration (EBUS-TBNA) has become the desired tool of choice for mediastinal and hilar lymph node staging and diagnosis.1–3 Due to advanced disease, a significant number of lung cancer patients are not primarily eligible for surgery; for them, only bronchoscopically obtained biopsies are available. With the evolution of personalized therapy for thoracic lung malignancies, sufficient tissue obtained by EBUS-TBNA is not only important for histopathological evaluation but also for the rapidly expanding number of molecular genetic tests. A multidirectional analysis of the EBUS-TBNA sample sufficient for molecular and pathologic assays following sample division has been presented by Nakajima et al.4 However, it is not known if dividing EBUS-TBNA samples for histopathological and molecular analysis can be implemented in routine day-to-day practice without having an impact on the histopathological and molecular assessments in lung cancer subjects.
Materials and methods
Patients
All patients presenting between October 2013 and March 2014 to our institution for EBUS-TBNA, with enlarged mediastinal or hilar lymphadenopathy detected by computed tomography (CT), were included in our study.
This study was approved by the Ethics Committee of University Duisburg-Essen, Essen, Germany (approval no 11-4921-BO) and informed consent was obtained from each patient.
EBUS-TBNA was performed in each lymph node measuring greater than 10 mm. In the event of negative or inconclusive histological results, further surgical procedures (mediastinoscopy or open thoracotomy) or a clinical follow-up within at least 6 months from surgery were performed. The lymph nodes from patients who underwent workup and were negative or who were inclusive for suspected lung cancer and did not show any evidence of progression of the adenopathy were considered negative (Figure 1).
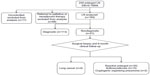  | Figure 1 Flowchart of the lymph nodes evaluated in the study (CONSORT). |
EBUS-TBNA
All procedures were performed with rigid bronchoscopy and jet ventilation under general anesthesia. The EBUS-TBNA examination was performed with an EBUS-TBNA bronchoscope (BF-UC 180F; Olympus Corporation, Tokyo, Japan) and all mediastinal and hilar lymph node stations were assessed systematically and punctured with a 22-gauge needle (NA-2015X-4022; Olympus Corporation) if their diameter exceeded 10 mm. After the needle was passed in the lymph node under ultrasound guidance, the internal stylet was retracted halfway and the needle was advanced back and forth to obtain further tissue aspirates. No additional vacuum was applied. The number of passes was decided by the bronchoscopist based on the amount of obtained biopsy material. No additional suction was applied.
Dividing the specimen for pathological and molecular analysis
The biopsy material was placed onto a glass slide and the internal stylet was advanced to facilitate removal of tissue from within. A drop of saline (0.9%) was added and a needle was used to mix and divide the liquid and solid material into two equivalent parts (Figure 2). One part was transferred to a tube, which contained 5 cc of formalin and then sent for histopathological evaluation. The other half was transferred into a 1.5 cc tube, which was immediately put on dry ice and stored at −80°C after the procedure. The remaining smeared material on the glass slide was sent for fast track cytological evaluation – rapid on-site cytology performed by the pathologist in a separate room. If the pathologist confirmed the first half as adequate for histopathological assessment, the second half was used for a new molecular test based on CK-19-mRNA concentration measurement.5,6 Otherwise, if more tissue was requested by the pathologist for diagnostic purposes, the second half was also used for histopathological assessment.7
RNA analysis
The samples were used to evaluate a new molecular test based on CK-19-mRNA concentration measurement. Eighty-five samples were centrifuged at 13,400 rpm for 5 minutes. The supernatant was discarded, 120 μL of Lynorhag (Sysmex, Tokyo, Japan) was added; afterwards, the sample was homogenized and centrifuged at 12,200 rpm for 60 seconds. Ten microliters of the lysate were used for CK-19-mRNA concentration measurement.
Statistical analysis
The sensitivity, specificity, and predictive values were calculated using the standard definitions. Statistical analysis was performed using SPSS software (v19.0; IBM, Armonk, NY, USA).
Results
Eighty-eight patients (66 male, 22 female; mean 66 years [range 49–87 years]) presenting with 249 enlarged mediastinal or hilar lymph nodes who met our inclusion criteria were enrolled in the study. All EBUS-TBNA samples were divided as described above. The punctured lymph node regions varied from one to seven (mean: 2.84) per patient and are listed in Table 1. Each lymph node region was punctured with one to six passes (mean: 3.18).
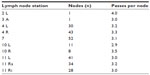  | Table 1 Overview of the lymph node stations sampled for histological and molecular analysis and the mean number of passes per node |
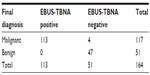
The cytological results confirmed malignant tissue in 113 cases (100 lung cancers: 41 adeno, 18 squamous cell, one adenosquamous, and 40 small-cell lung cancer [SCLC], plus 13 other cancers: nine colorectal, two Hodgkin’s disease, and two squamous cell carcinoma of the hypopharynx).
Out of 136 benign or ambiguous results, 51 were followed up (nine mediastinoscopy, 19 thoracotomy, two thoracoscopy, and 21 CT/positron emission tomography [PET]-CT). In one case, histopathological evaluation of the whole lymph node revealed malignant tissue (adenosquamous carcinoma); in three cases PET-CT revealed hypermetabolic activity consistent with malignant disease; the other 47 were verified to be benign (15 anthracosilicosis, 30 reactive enlarged, and two cryptogenic organizing pneumonia).
The remaining 85 cases were not reevaluated by surgical means. In 68 cases, cytological evaluation had revealed malignant tissue in a different lymph node station and these patients received neoadjuvant or palliative treatment. Another 17 were lost to follow-up. The sensitivity, specificity, negative predictive value, and accuracy of EBUS-TBNA for detecting malignancy were 96.6%, 100%, 92.2%, and 97.6%, respectively (Table 2).
In six cases, the pathologist required the second half of the EBUS-TBNA specimen for further histopathological evaluation. This did not lead to a change of the histopathological result in any of the cases.
Eighty-five of the second half samples (35 SCLC, 26 adenocarcinoma, 15 squamous cell carcinoma, nine no proof of malignant cells) were sent for a new molecular test based on cytokeratin-19-mRNA concentration measurement.5,6 Out of these, 63 (74.1%) could be successfully performed.
Discussion
Our study shows that dividing EBUS-TBNA specimens and preserving a part of the sample for molecular analysis (ie, CK-19-mRNA concentration measurement, etc) can be implemented in routine practice without having a negative effect on histopathological results.
Assessing tumor tissue for genetic variants such as in the epidermal growth factor receptor (EGFR) and the Kirsten rat sarcoma viral oncogene homolog (KRAS) genes, and the echinoderm microtubule-associated protein-like 4 – anaplastic lymphoma kinase fusion gene (EML4-ALK), has become the standard of care, especially in support of personalized chemotherapy treatments for lung cancer patients.8 Other predictive or diagnostic biomarkers such as v-raf murine sarcoma viral oncogene homolog B (BRAF) mutations or short stature homeobox 2 (SHOX2) hypermethylation have been proven to be helpful in experimental studies.9–11 With the rapidly increasing number of molecular markers, it is pivotal to ensure that we are set to provide not only sufficient material but also tissue processed by the best and most efficient means for optimal patient treatment.
Nowadays, lung cancer diagnosis and staging is achieved with minimally invasive means, in particular with EBUS-TBNA, especially in the case of advanced stage tumors. Therefore, only small biopsies are provided for histopathological evaluation, immunostaining, and a growing number of molecular analyses.12,13 Assays for EGFR, KRAS, and EML4-ALK can be applied on a formalin-fixed paraffin-embedded (FFPE) EBUS-TBNA specimen.14–16 Moreover, EBUS-TBNA provides high-quality RNA for biomarker testing.17 However, there is doubt as to whether upcoming methods such as miRNA profiling can be performed on FFPE material.18–20 Alternative methods such as cryopreservation with liquid nitrogen or the use of RNA preservative solutions are cost-intensive. A standardized extraction of DNA and RNA from such specimens that is easy to apply will become a key factor in the era of molecularly targeted personalized therapy.
Not every EBUS-TBNA needle pass contains tumor cells. Lee et al showed that the optimal diagnostic yield can be achieved after three needle passes.21 If specimens are being sent to different laboratories or are intended to be analyzed by different methods, one has to be sure that every sample contains sufficient tumor material to enable adequate analysis. We previously conducted a study obtaining three separate EBUS-TBNA passages for molecular analysis in addition to the standard diagnostic procedure.10 The proposed method in this study is less time-consuming and more cost-efficient in everyday practice. Sufficient amounts of tumor cells for both diagnosis and further molecular analysis can be provided. Developing a method that achieves molecular profiling in low-volume samples containing a known percentage of tumor cells is of paramount clinical importance.
It is known that SCLC does not always express CK-19-mRNA. The success rate of molecular testing performed on EBUS-TBNA samples would probably have been higher if additional tests had been performed.22
Our results demonstrate the feasibility of dividing EBUS-TBNA specimens without loss of accuracy in histopathological and molecular analyses. This allows clinicians to perform RNA profiling and tissue diagnosis on EBUS-TBNA material without the need for extra biopsies and with secure tumor cell content.
Acknowledgments
The authors wish to thank Heiko Hang, Rüdiger Karpf-Wissel, and Maria Radner (Ruhrlandklinik Essen, Germany) for their assistance in data collection. All of the authors contributed significantly to the manuscript and made substantial contributions to one or more of the conception and design, acquisition of data, analysis and interpretation of data, drafting of the submitted article, critical revision for important intellectual content, and final approval of the version to be published. KD serves as a guarantor of the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
Varela-Lema L, Fernández-Villar A, Ruano-Ravina A. Effectiveness and safety of endobronchial ultrasound-transbronchial needle aspiration: a systematic review. Eur Respir J. 2009;33(5):1156–1164. | |
Herth FJ, Eberhardt R, Vilmann P, Krasnik M, Ernst Al. Real-time endobronchial ultrasound guided transbronchial needle aspiration for sampling mediastinal lymph nodes. Thorax. 2006;61(9):795–798. | |
Yasufuku K, Pierre A, Darling G, et al. A prospective controlled trial of endobronchial ultrasound-guided transbronchial needle aspiration compared with mediastinoscopy for mediastinal lymph node staging of lung cancer. J Thorac Cardiovasc Surg. 2011;142(6):1393–1400. e1. | |
Nakajima T, Yasufuku K. How I do it – optimal methodology for multidirectional analysis of endobronchial ultrasound-guided transbronchial needle aspiration samples. J Thorac Oncol. 2011;6(1):203–206. | |
Wallace MB, Block MI, Gillanders W, et al. Accurate molecular detection of non-small cell lung cancer metastases in mediastinal lymph nodes sampled by endoscopic ultrasound-guided needle aspiration. Chest. 2005;127(2):430–437. | |
Bugalho A, Martins C, Dias SS, et al. Cytokeratin 19, carcinoembryonic antigen, and epithelial cell adhesion molecule detect lung cancer lymph node metastasis in endobronchial ultrasound-guided tranbronchial aspiration samples. Clin Lung Cancer. 2013;14(6):704–712. | |
Lawson MH, Rassl DM, Cummings NM, et al. Tissue banking of diagnostic lung cancer biopsies for extraction of high quality RNA. J Thorac Oncol. 2010;5(7):956–963. | |
Nana-Sinkam SP, Powell CA. Molecular biology of lung cancer: Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest. 2013;143(Suppl 5):e30S–e39S. | |
Wiesweg M, Ting S, Reis H, et al. Feasibility of preemptive biomarker profiling for personalised early clinical drug development at a Comprehensive Cancer Center. Eur J Cancer. 2013;49(15):3076–3082. | |
Darwiche K, Zarogoulidis P, Baehner K, et al. Assessment of SHOX2 methylation in EBUS-TBNA specimen improves accuracy in lung cancer staging. Ann Oncol. 2013;24(11):2866–2870. | |
Brustugun OT, Khattak AM, Trømborg AK, et al. BRAF-mutations in non-small cell lung cancer. Lung Cancer. 2014;84(1):36–38. | |
Scarpa A, Sikora K, Fassan M, et al. Molecular typing of lung adenocarcinoma on cytological samples using a multigene next generation sequencing panel. PLoS One. 2013;8(11):e80478. | |
Herbst RS, Lippman SM. Molecular signatures of lung cancer – toward personalized therapy. N Engl J Med. 2007;356(1):76–78. | |
Santis G, Angell R, Nickless G, et al. Screening for EGFR and KRAS mutations in endobronchial ultrasound derived transbronchial needle aspirates in non-small cell lung cancer using COLD-PCR. PLoS One. 2011;6(9):e25191. | |
Nakajima T, Kimura H, Takeuchi K, et al. Treatment of lung cancer with an ALK inhibitor after EML4-ALK fusion gene detection using endobronchial ultrasound-guided transbronchial needle aspiration. J Thorac Oncol. 2010;5(12):2041–2043. | |
Garcia-Olivé I, Monsó E, Andreo F, et al. Endobronchial ultrasound-guided transbronchial needle aspiration for identifying EGFR mutations. Eur Respir J. 2010;35(2):391–395. | |
Schmid-Bindert G, Wang Y, Jiang H, et al. EBUS-TBNA provides highest RNA yield for multiple biomarker testing from routinely obtained small biopsies in non-small cell lung cancer patients – a comparative study of three different minimal invasive sampling methods. PLoS One. 2013;8(10):e77948. | |
Kojima K, April C, Canasto-Chibuque C, et al. Transcriptome profiling of archived sectioned formalin-fixed paraffin-embedded (AS-FFPE) tissue for disease classification. PLoS One. 2014;9(1):e86961. | |
Fedorowicz G, Guerrero S, Wu TD, Modrusan Z. Microarray analysis of RNA extracted from formalin-fixed, paraffin-embedded and matched fresh-frozen ovarian adenocarcinomas. BMC Med Genomics. 2009;2:23. | |
Freidin MB, Bhudia N, Lim E, Nicholson AG, Cookson WO, Moffatt MF. Impact of collection and storage of lung tumor tissue on whole genome expression profiling. J Mol Diagn. 2012;14(2):140–148. | |
Lee HS, Lee GK, Lee HS, et al. Real-time endobronchial ultrasound-guided transbronchial needle aspiration in mediastinal staging of non-small cell lung cancer: how many aspirations per target lymph node station? Chest. 2008;134(2):368–374. | |
Kanaji N, Bandoh S, Ishii T, et al. Cytokeratins negatively regulate the invasive potential of lung cancer cell lines. Oncol Rep. 2011;26(4):763–768. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.