Back to Journals » International Journal of Nanomedicine » Volume 17
Multi-Layered PLGA-PEI Nanoparticles Functionalized with TKD Peptide for Targeted Delivery of Pep5 to Breast Tumor Cells and Spheroids
Authors Mohan AK , M M, Kumar TRS, Kumar GSV
Received 7 June 2022
Accepted for publication 5 October 2022
Published 22 November 2022 Volume 2022:17 Pages 5581—5600
DOI https://doi.org/10.2147/IJN.S376358
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Anderson Oliveira Lobo
Akhil K Mohan,1,2 Minsa M,3 T R Santhosh Kumar,3 G S Vinod Kumar1
1Nano Drug Delivery Systems (NDDS), Cancer Biology Division, Rajiv Gandhi Centre for Biotechnology, Thiruvananthapuram, Kerala, 695014, India; 2Research Centre, Department of Biotechnology, University of Kerala, Thiruvananthapuram, Kerala, India; 3Cancer Research Programme-1, Bio-Innovation Center (BIC), Rajiv Gandhi Centre for Biotechnology, Thiruvananthapuram, Kerala, 695014, India
Correspondence: G S Vinod Kumar, Tel +91 471 2781217, Fax +91 471 2348096, Email [email protected]
Purpose: Peptide-based therapy is a promising strategy for cancer treatment because of its low drug resistance. However, the major challenge is their inability to target cancer cells specifically. So, a targeted nano-delivery system that could deliver therapeutic peptides selectively to cancer cells to stimulate their action is highly desirable. This study aims to deliver the antitumor peptide, Pep5, to breast tumor cells selectively using a targeting peptide functionalised multi-layered PLGA-PEI nanoparticles.
Methods: In this study, Pep5 entrapped PLGA-PEI (Pep5-PPN) dual layered nanoparticles were developed. These nanoparticles were decorated with TKD (Pep5-TPPN) on their surface for site-specific delivery of Pep5 to breast tumor cells. The particles were then characterized using various instrumental analyses. In vitro cytotoxicity of the particles was evaluated in estrogen receptor positive (ER+ve) and triple negative breast cancer (TNBC) cells. An ex vivo tumor spheroid model was used to analyze the antitumor activity of the particles.
Results: Uniformly round Pep5-TPPN particles were synthesized with an average diameter of 420.8 ± 14.72 nm. The conjugation of PEI over Pep5-PLGA nanoparticles shifted the zeta potential from − 11.6 ± 2.16 mV to +20.01 ± 2.97 mV. In vitro cytotoxicity analysis proved that TKD conjugation to nanoparticles enhanced the antitumor activity of Pep5 in tested breast cancer cells. Pep5-TPPN induced cytoskeletal damage and apoptosis in the tested cells, which showed that the mechanism of action of Pep5 is conserved but potentiated. Active targeting of Pep5 suppressed the tumor growth in ex vivo spheroid models.
Conclusion: A multi-layered nanoparticle functionalized with dual peptide was fabricated for active tumor targeting, which stimulated Pep5 activity to reduce the tumor growth in vitro and ex vivo.
Keywords: peptide, Pep 5, tumor, cancer, breast
Graphical Abstract:
Introduction
Breast cancer is the most commonly diagnosed malignancy accounting for more than 24% of all cancer incidence and the leading cause of cancer-related deaths in females, around 15%, worldwide.1 It is often treated with surgery, chemotherapy, radiation, and targeted therapy.2 Among these strategies, targeted therapy based on peptides has been of great interest because of their unique benefits, including low molecular weight, ability to specifically target tumour cells, and minimal toxicity in normal tissues.3
Peptide 5 (pep5; WELVVLGKL) is a natural intracellular peptide derived by the degradation of Cyclin D2 through the ubiquitin–proteasome system. It was first identified in the extracts of HeLa cells, which specifically increased during the S phase of the cell cycle.4–6 This peptide induced cell death when introduced into MDA-MB-231 breast cancer cells, which expressed low levels of Cyclin D2.6 Pep5 promotes a substantial cytoskeletal disruption most likely by binding to Plectin and/or CLIC1, which appears to contribute to cell death pathways.6 These studies show the potential therapeutic capabilities of pep5, making it a candidate for treating breast cancer. However, instability, non-targeting and short plasma half-life are the major drawbacks encountered by the peptide-based therapy.7 To overcome these drawbacks, it is important to introduce biocompatible targeted nanocarriers capable of efficiently delivering the peptide at the target site.
With the advent of nano-biotechnology, there is an unprecedented increase in the design and synthesis of polymer-based nanocarriers for cancer treatment. The type of polymer and the method of synthesis allows for manipulation of the properties of the carrier molecules, such as biodistribution and bioavailability of the drug target.8 Biopolymers like poly(lactic-co-glycolic acid) (PLGA) are frequently used for constructing anticancer drug delivery nanocarriers.9 This FDA-approved polymer is well known for its properties, like biodegradability, biocompatibility, enhanced protection of the drug from degradation, and the possibility of sustained-release.10,11 These unique features make them attractive for constructing targeted drug delivery systems. However, therapeutic nanoparticles have certain disadvantages. The major disadvantage is the interaction with nonspecific cells and proteins, resulting in the accumulation of drugs in non-target tissues.12 Hence, proper targeting is required to efficiently induce the pharmacological action of the loaded drug without damaging healthy tissues. Active targeting molecules like ligands and antibodies have been conjugated to the nanocarriers to direct them towards the overexpressed receptors on the tumor cells, thereby improving the targetability.13 However, their application is limited in breast cancer therapy due to the heterogeneity in the expression of different receptors.
Unlike the normal cells, the tumor cells including lung cancer cells, colorectal cancer cells, leukemic cells, and breast cancer cells express heat-shock protein 70 on their plasma membrane (memHsp70), which is also up-regulated in the metastatic tumors.14–17 It is reported that these proteins are overexpressed in Triple-Negative Breast Cancer cells also.14 These features make memHsp70 a potential target for tumor diagnostic and therapeutic procedures. A 14-mer peptide (TKD: CTKDNNLLGRFELSG) derived from the oligomerization domain of memHsp demonstrated selective binding and rapid internalization in different cancer cells with a memHsp70-positive phenotype in vitro.18 The in vivo targeting ability of the peptide was also proved in breast cancer mice models after coating the nanoparticles with TKD peptide.19 Hence, the conjugation of TKD peptide on the surface of nanocarriers represents a means of their tumor homing irrespective of the different expression statuses of growth factor receptors.
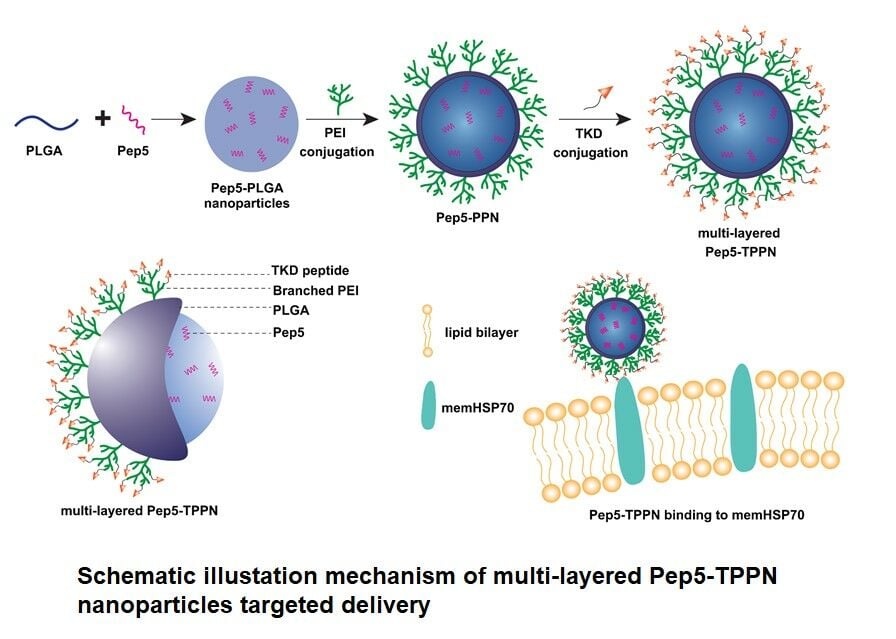
In the present study, a targeted multi-layered nanocarrier system loaded with the cytotoxic peptide-Pep5 for their stimulated activity against breast cancer was developed. The system is composed of Pep-5 entrapped PLGA nanoparticles coated on the surface with branched PEI (bPEI) via an amide bond (Scheme 1A). Besides promoting endosomal escape, PEI coating allows reversing the surface potential of PLGA nanoparticles from negative to positive, which allows the nanoparticle to bind to negatively charged membrane of the cells and be retained in the tumor tissue.20,21 TKD peptide was layered over PEI coating to obtain a multi-layered active targeting system to ER+ve and TNBC cells (Scheme 1B). The synthesized nanoparticles were subjected to a series of physicochemical characterisation. To address the enhanced antitumor activity of Pep5 and to check whether Pep5-TPPN could potentially be used for breast cancer therapy, cytotoxicity, live/dead, clonogenic and cell uptake assays were performed. Pep5-TPPN induced activation of apoptosis was carefully investigated by cleaved Caspase-3 immunofluorescence and Western blotting. Moreover, tumor targeting ability of Pep5-TPPN was evaluated in an ex vivo tumor spheroid model. To our knowledge, this study presents the first attempt to stimulate the antitumor activity of Pep5 with actively targeted nanoparticles to breast cancer cells.
 |
Scheme 1 Schematic illustation mechanism of bPEI conjugation to PLGA and its schematic represenataion. |
Materials and Methods
Materials
Fmoc-protected amino acids and TentaGel TM –S–NH2 were purchased from Peptide International Inc. HMPA linker was purchased from Novabiochem. Coupling reagents, hydrobenzoxy triazole (HOBT), diisopropyl ethylamine (DIEA), and O-benzotriazole-N,N,N’,N’-tetramethyl-uronium-hexafluoro-phosphate (HBTU), and cleavage reagents, trifluoroacetic acid (TFA) and triisopropylsilane (TIS), were obtained from Merck. Piperidine, ninhydrin, dichloromethane (DCM), diethyl ether, N,N-dimethyl formamide (DMF) were obtained from Spectrochem. PLGA, branched PEI (bPEI), PVA, MTT, 1-(3-dimethylaminopropyl)-3-3-ethylcarbodiimide hydrochloride (EDC), N-hydroxysuccinimide (NHS), and all other chemicals were purchased from Sigma-Aldrich (Steinheim, Germany). Fetal bovine serum (FBS) was purchased from Gibco (Life Technologies AG, Basel, Switzerland). DMEM, Live Dead Assay Kit, and Rhodamine-Phalloidin were purchased from Invitrogen. Breast cancer cells MDA-MB-231, MCF7, and MDA-MB-231 cells expressing plasma membrane-targeted GFP were procured from the cell repository, RGCB. All cells were cultured in DMEM medium containing 10% FBS and 1% antibiotics.
Synthesis and Characterization of Peptides
Cytotoxic peptide (Pep5: WELVVLGKL) and targeting peptide (TKD: CTKDNNLLGRFELSG) were synthesized using the standard F-moc method and purified by high-performance liquid chromatography (HPLC). The molecular mass of the synthesized peptide was analyzed by mass spectrometry using MALDI-TOF mass spectrometer.
In vitro Cytotoxicity Study of Pep5
Cytotoxicity of Pep5 was examined on HaCaT, MCF7 and MDA-MB-231 using methyl thiazolyl diphenyl tetrazolium bromide (MTT) assay. This method for measuring cytotoxicity is based on the ability of viable cells to cleave the tetrazolium compound by mitochondrial dehydrogenase. Briefly, cells were seeded in 96-well culture plates at a cell density of 5 × 103 cells/well in DMEM cell culture medium and incubated overnight for cells to attach to the wells. These cells were treated with different concentrations of Pep5, ranging from 10 to 150 µM. Cell lines with no treatment served as controls. After 48h of treatment, 100 µL of MTT reagent solution (5 µg/mL) was added to the cells and incubated for 4h at 37℃. Absorbance was recorded at 570nm using a Microplate Reader after dissolving the developed purple formazan crystals with 200 µL IPA. Data were plotted using cytotoxicity percentage against the concentration of treatment. All the experiments were performed in triplicates. Cytotoxicity percentage was calculated using the formula:
Viability (%) = (OD of sample/OD of control) x 100
Cytotoxicity (%) = 100 – Viability
Synthesis of Pep5 Entrapped TKD Conjugated PLGA-PEI (Pep5-PPN) Nanoparticles
Synthesis of Pep5 Entrapped PLGA Nanoparticles
The Cytotoxic peptide, Pep5, was encapsulated in PLGA nanoparticles by a modified double emulsion solvent evaporation method.22 In brief, 100mg of PLGA dissolved in 4mL of dichloromethane was emulsified under sonication (30s, 20W) over an ice bath with a solution containing 4 mg Pep5. This first emulsion was added dropwise to 30 mL of an aqueous surfactant solution (1% PVA) with constant stirring, and the mixture was emulsified again by sonication (30s, 20W), creating a double emulsion. The solvent was then allowed to evaporate by stirring the emulsion at 600 rpm overnight at room temperature. The particles were then collected by centrifugation at 12,000 rpm for 30 min at 4℃. The pellet was washed three times and finally re-suspended in deionized water and then lyophilized. Rhodamine-B entrapped PLGA particles, and bare PLGA particles without Pep5 were prepared using the same protocol.
Chemical Conjugation of Polyethyleneimine to PLGA Nanoparticles
About 0.1M solution of N-hydroxysuccinimide (NHS) and 1-(3-dimethyl aminopropyl)-3-3-ethylcarbodiimide hydrochloride (EDC) in PBS (pH 7.4) was used for activating the carboxyl groups on the PLGA nanoparticles. Two milliliters of each solution was added to the PLGA nanoparticle suspension in MES buffer and stirred for 4h at room temperature. Activated PLGA nanoparticles were further sonicated using a Vibra CellTM Probe Sonicator for 2 min. About 250 µL of bPEI suspension was added to the nanoparticle suspension and stirred for 12h at room temperature. Finally, PEI-conjugated PLGA nanoparticles were harvested by centrifugation at 12,000 rpm for 30 min at 4℃. After washing three times, the particles were suspended in distilled water, lyophilized, and stored at 4℃ until used.23 Rhodamine-B entrapped PLGA particles and bare PLGA nanoparticles were also coated with PEI using the same method to give Rhodamine-B-PLGA-PEI nanoparticles (R-PPN) and and B-PLGA-PEI nanoparticles (B-PPN).
Targeting Peptide (TKD) Conjugation to Pep5-PPN
TKD (2mg) peptide was coupled to Pep5-PPN by EDC-NHS method. For that, an aqueous solution of TKD (1 mg/mL), with a constant stirring, was added with 0.5 mL of EDC (1mM) and 0.5 mL NHS (1mM) at room temperature. The mixture is allowed to stir at room temperature for 4 hours. The activated TKD was then added to Pep5-PPN solution dropwise at a rate of 1 mL/min at room temperature. After stirring overnight, the TKD conjugated Pep5-PPN (Pep5-TPPN) was obtained through centrifugation at 12,000 rpm for 30 minutes at 4℃. The pellet was washed and re-suspended in distilled water and lyophilized. R-PPN and B-PPN were also conjugated with TKD using the same protocol to yield R-TPPN and B-TPPN.
Characterization
FTIR Spectroscopy
The effective conjugation of PEI with PLGA nanoparticles was confirmed by analysing Fourier Transform Infrared (FT-IR) spectra of lyophilised PPN using FT-IR spectrometer (Spectrum 65, Perkin-Elmer, USA). FTIR spectrum of PPN obtained with potassium bromide (KBr) pellet was compared with that of PLGA nanoparticles and PEI.
Differential Scanning Calorimetry
Differential scanning calorimetry (DSC) thermograms of PPN nanoparticles, PLGA nanoparticles and PEI were analyzed. DSC thermograms were obtained using an automatic thermal analyzer system (Pyres DSC, Perkin-Elmer, USA). Samples were sealed in standard aluminium pans and heated from 0℃ to 250℃ at a heating rate of 10℃/min. Nitrogen was purged into the system constantly at a rate of 20 mL/min. An empty pan, sealed in the same way as the sample, was used as a reference.
1H NMR
The NMR spectrum of PPN was obtained with Bruker Advance 400 MHz NMR spectrometer (Hannover, Germany) using DMSO as the solvent.
Particle Size Analysis
The size distribution of the synthesized nanoparticles was analyzed using the dynamic light scattering method (DLS), a Delsa TM particle size analyzer (Beckman Coulter Nano Particle Analyzer).
Zeta Potential Measurements
Zeta potential measurement was carried out to find the surface charge of the nanoparticles using Zetasizer Nano ZS (Malvern Instruments, Malvern, UK).
TEM Analysis
The size and morphology of different nanoparticles synthesized were also analyzed using a transmission electron microscope (TEM, JEOL 1011, and Japan). For TEM, the nanoparticle samples were prepared by suspending them in water milli-Q at 25℃. The samples were then dropped onto copper TEM grids, and measurements were taken after the samples were completely dried in a desiccator.
SEM Analysis
Morphological analysis of Pep5-PLGA nanoparticles and Pep5-TPPN was further carried out using field emission scanning electron microscopy (FE-SEM) (FEI Nova NanoSEM 450).
Yield, Peptide Loading Percentage and Encapsulation Efficiency Calculation
The amount of Pep5 encapsulated in the weighed amount of PLGA nanoparticles was evaluated by measuring the UV absorbance using a UV spectrophotometer (Perkin-Elmer, USA) at 280 nm. The absorbance was compared with a standard curve of Pep5 to obtain the amount of Pep5 encapsulated in the nanoparticle. Yield, peptide loading percentage and encapsulation efficiency were calculated using the following formulae
Yield = weight of nanoparticle/(initial weight of polymer + initial weight of Pep5) x 100
Peptide loading percentage = (weight of peptide encapsulated/weight of nanoparticle) x 100
Encapsulation efficiency = (weight of peptide encapsulated/initial weight of peptide) x 100
In vitro Peptide Release
The in vitro release study of Pep5-PPN nanoparticles was operated in triplicate. The study was performed by incubating 10mg of nanoparticles in 5mL of 1 M phosphate-buffered saline (pH 6.2 and 7.4) in a dialysis bag and dialyzed against 50 mL PBS taken in a glass beaker. The nanoparticle suspensions were continuously stirred in a thermo-shaker (120 rpm) at 37℃. At preselected times, in triplicate, 1 mL of sample was collected, and optical density was measured at 280 nm in a spectrophotometer. Every time, the removed supernatant was replaced by fresh PBS.
Intracellular Uptake Studies
To evaluate the intracellular uptake of the synthesized nanoparticles, confocal laser scanning microscope (CLSM) and flow cytometry analysis were performed. Briefly, for CLSM, MDA-MB-231 cells stably expressing plasma membrane-targeted EGFP were grown in coverslips kept on microwell dishes at a seeding density of 1 × 105 cells per coverslip overnight. Afterwards, the medium was replaced with a suspension of R-PPN and R-TPPN and cells were then incubated at 37°C for 4hrs. Nuclei were counterstained with Hoechst. Prior to imaging, cells were washed with PBS to remove un-internalised nanoparticles. The images were acquired using a confocal laser scanning microscope (CLSM) (TCS SP2; Leica Microsystems, Germany). Cell internalisation was also checked in MCF7 cells, where nuclei were counterstained with Hoechst and cell membrane using CellMask TM (Thermo Scientific) green plasma membrane stain. For flow cytometry analysis, 1 × 105 cells (both MCF7 and MDA-MB-231 cells) grown on a 24 well plate were incubated with R-PPN and R-TPPN for 4 hours. The cells were trypsinized, washed with PBS and subjected to flow cytometry (BD FACSAria III, USA) analysis.
Cytotoxicity Analysis by MTT
Briefly, MCF7 and MDA-MB-231 cells were seeded in a 96-well plate at a density of 5 × 103 cells/well and incubated overnight to attach to the wells. Cells were treated with 100 µmol/L Pep5, Pep5-PLGA-PEI, and TKD-Pep5-PLGA-PEI. Cells that received no treatment were served as control. At predetermined time points (24, 48, and 72 hr) of treatment, MTT reagent solution (100 µL of 5 mg/mL MTT in DMEM) was added to the cells and incubated for 2hrs at 37°C. Absorbance reading was recorded at 570 nm using a Microplate Reader (iMark™ Biorad) after dissolving the developed purple formazan crystals with isopropanol. Data were plotted using cytotoxicity percentage against the concentration of treatment. All experiments were performed in triplicates.
Cytotoxicity percentage was calculated using the formula:
Viability (%) = (OD of sample/OD of control) x 100
Cytotoxicity (%) = 100 – Viability
Clonogenic Assay
The clonogenic assay is a cell survival study based on the potential of a single cell to develop into a colony following treatment. This study was performed to see how the Pep5-NPs affected the clonogenic potential of breast cancer cells. For this, MCF7 and MDA-MB-231 cells were seeded (4x103 cells/well) in a 96-well plate and allowed to attach by overnight incubation at 37℃. Then, the cells were treated with Pep5, Pep5-PPN and Pep5-TPPN (at Pep5 concentration 100 µM). After 48 hours of treatment, cells were collected by trypsinisation and re-seeded on 60mm plates. These cells were allowed to form colonies for ten days with regular media changes on each day. Finally, visible colonies were fixed using 4% PFA and stained with methylene blue for 20 minutes. The clonogenicity index was calculated using the formula,
Clonogenicity index = (Number of colonies in treatment/number of colonies in control)* 100
Live Dead Assay
The live-dead assay was performed to study the efficacy of Pep5 and Pep5-NPs in imparting cytotoxicity on MCF7 and MDA-MB-231 cells. CLSM (TCS SP2; Leica Microsystems, Germany) and flow cytometry (BD FACSAria III, USA) analysis were adapted for visualisation and quantification of cell death, respectively. The experiment was carried out based on the manufacturer’s protocols. After treating the cells with Pep5, Pep5-PPN and Pep5-TPPN (at Pep5 concentration 100 µM) for 48 hours, live cells were stained with calcein (excitation/emission spectra at 494/517 nm) and dead cells with ethidium homodimer (excitation/emission spectra at 528/617 nm) for 15 minutes in the dark.
Lysosomal Integrity Assessment Using Acridine Orange Staining
To assess the impact of Pep5-NPs on lysosome, acridine orange staining was performed. Briefly, 1 × 105 cells were seeded into a 12 well plate and incubated overnight for cell attachment. The attached cells were then treated with Pep5, Pep5-PPN and Pep5-TPPN formulations for 48 hours. After that, cells were incubated with acridine orange (4µg/mL) for 15 min at 37℃. Prior to imaging, cells were washed with PBS. Stained cells were imaged using fluorescent microscope (Olympus IX73) under green and red filters to visualise the integrity of lysosomes.
Rhodamine Phalloidin Staining
Rhodamine Phalloidin staining, which specifically stains the actin filaments, was performed on MCF7 and MDA-MB-231 cells to examine the influence of Pep5 loaded formulations on the cytoskeletal organisation of the cells.24 Briefly, 2 × 104 cells were seeded per chamber on 8 chamber cell imaging coverglass and incubated at 37℃ overnight under 5% CO2. Media was then removed, and cells were exposed to Pep5, Pep5-PPN and Pep5-TPPN. Further 48 hours of incubation, cells were fixed using 3.7% paraformaldehyde for 10 min. Post-fixing, cells were permeabilized with 0.1% triton-X 100 for 2 min and then blocked using 1% BSA for 20 min at room temperature. Finally, cells were stained with Rhodamine Phalloidin for 20 min and counterstained with Hoechst 33,342 for 5 min prior to confocal imaging (For MCF7 cells: A1R, Nikon, Japan & for MDA-MB-231 cells: TCS SP2; Leica Microsystems, Germany) for morphology analysis.
Chromosome Condensation Assay
For the primary evaluation of apoptosis by detecting nuclear condensation, DAPI staining was performed in MCF7 and MDA-MB 231 cell lines. Cells were grown on 8 chamber cell imaging coverglass at a density of 2 × 104 cell/chamber and incubated at 37℃ with 5% CO2. After incubation, cells were exposed to Pep5, Pep5-PPN and Pep5-TPPN (at Pep5 concentration 100 uM) for 48 hours. Subsequently, cells were washed with PBS and fixed in 4% PFA for 10 min at room temperature prior to DAPI staining for 5 min. Finally, the stained chromosomes were visualised using a confocal laser scanning microscope (For MCF7 cells: A1R, Nikon, Japan & for MDA-MB-231 cells: TCS SP2; Leica Microsystems, Germany).
Analysis of Apoptosis by Immunofluorescence for Caspase-3
To examine whether Pep-5 and Pep5-NP induces apoptosis by cleaving Caspase-3, an immunofluorescence study was performed. MCF-7 and MDA-MB-231 cells were grown in 8 chamber coverglass (2x104 cells/chamber) at 37℃. Media was then replaced with different treatment samples, including Pep5, Pep5-PPN and Pep5-TPPN. After 48 h incubation, cells were washed thrice with PBS and fixed with ice-cold 4% paraformaldehyde at 4℃ for 10 min. Cells were further blocked using 10% normal goat serum containing 0.1% Triton-X 100 for 45 mins at room temperature. Cleaved Caspase-3 primary antibody (Cell signaling technologies, #9664) was then introduced to each well and incubated overnight at 4℃. Following incubation, cells were added with anti-rabbit secondary antibody tagged with Alexa fluor 488 (Abcam, #150077) for 1 h. At the end of incubation, nucleus was stained with Hoechst 33,342 for 5 mins. Finally, the cells were imaged using confocal laser scanning microscope (For MCF7 cells: A1R, Nikon, Japan & for MDA-MB-231 cells: TCS SP2; Leica Microsystems, Germany).
Apoptotic Index was calculated by the formula:
Apoptotic Index = (Cleaved caspase 3 positive cells/Total number of cells) x 100
Western Blotting
Western blot analysis was carried out to further confirm the potential of Pep5 and Pep5-NPs to induce apoptosis. Briefly, after being treated with Pep5 and Pep5-NPs for 48 hours, MDA-MB-231 cells were lysed in lysis buffer (Qproteome Mammalian Protein Prep Kit, Qiagen), and the total protein extracted was quantified using Bradford’s method. An aliquot of 30 µg of total protein was resolved in an SDS-PAGE and immunoblotted with specific antibodies, including PARP (Cell signalling technology, #9542S), Caspase-3 (Cell signalling technology, #9662S) and β-actin (Sigma Aldrich, #A5316). The protein bands were obtained under a bioanalytical imaging system (Azure Biosystems, Inc.) using a chemiluminescent substrate (Supersignal West Pico PLUS, Thermo Scientific).
Ex vivo Anti-Cancer Therapeutic Efficacy
For culturing spheroids, 96-well plates were precoated with 1.5% agarose and sterilized by UV exposure. Cells grown as a monolayer were harvested by trypsinisation, and the cell number was counted using a hemocytometer. 1×103 cells were seeded per well in DMEM medium supplemented with 10% FBS. After 3 days, tumor spheroids of uniform diameter were selected and treated with different formulations (free Pep5, B-PPN, Pep5-PPN and Pep5-TPPN) in a fresh medium for 48 hours, along with untreated spheroids being the negative control. The spheroids were imaged at 0, 24 and 48 h time points and diameter was calculated with a computer assisted analysis system (CellSens, Olympus). After 48 hours of treatment, a live dead assay was done on the treated spheroids using Calcein and Ethidium homodimer, which stains live and dead cells with green and red fluorescence, respectively. The fluorescent images were captured in a fluorescent microscope (Olympus) under 10X magnification.
Statistics
All the experiments were performed in triplicate. Statistical analysis of the data was done by performing one way, and two-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison tests using GraphPad Prism v8.0.2. (GraphPad Software Inc., San Diego, CA, USA). The results were expressed as an arithmetic mean ± standard deviation of the mean, and the differences were considered significant when P < 0.05 (*), p < 0.01 (**), p < 0.001 (***) or p < 0.0001 (****).
Results and Discussions
Synthesis and Characterization of Peptides
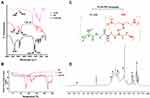
Peptides, TKD, and Pep5 were synthesized using the solid-phase peptide synthesis technique using F-moc chemistry with good purity and yield. The molecular weight of the peptides was investigated by mass spectrometry using MALDI-TOF mass spectrometer and is found to be 1667.38 and 1056.62 (Figure 1A and B), which agrees with the calculated values 1666.85 and 1056.29. This figure also shows the absence of other peaks, which indicates that no amino acid was added or deleted to the original sequence of the peptide during the synthesis process.
In vitro Cytotoxicity of Peptide
In vitro cytotoxicity of the Pep5 on breast cancer cells was analyzed by MTT assay using a peptide concentration ranging from 10μM to 150μM, and they exerted significant toxicity on the tested cell lines at 48 hours. Cytotoxicity is increased in a dose-dependent manner in MCF7 and MDA-MB-231 cells. This increase in cytotoxicity demonstrates the bioactivity of the peptide. Pep5 at a concentration of 100μM showed considerable cytotoxicity of 44.97 ± 0.47% and 40.2 ± 0.54% on MCF7 and MDA MB 231 cells, respectively. The higher concentration of peptides (>100 μM) showed no significant increase in cytotoxicity (Figure 1C and D). So Pep5 concentration of 100 μM was used for further analysis. Interestingly, HaCaT cells were not significantly affected at all the tested concentrations (Figure 1E), which proves the biocompatibility of the peptide in cells that express normal level of Cyclin D2. With this selectivity to tumor cells, Pep5 exhibit superior cytotoxicity as with already described anticancer peptides.25–29
Synthesis of Pep5-PPN
Pep5 was encapsulated in PLGA nanoparticles by a modified water/oil/water double emulsion solvent evaporation method. PEI was then conjugated to the surface of PLGA nanoparticle by the EDC-NHS method. The formation of the amide bond between PLGA and PEI was confirmed by FTIR spectroscopy. In (Figure 2A), the peak at 3000 cm−1 in PLGA and 2947 cm−1 in PEI can be attributed to the aliphatic –C–H– stretching present in these polymers. The presence of methylenic –C–H– stretching is indicated by the peaks present at 2949 cm−1 and 2819 cm−1 in the IR spectra of PLGA and PEI, respectively. The peak at 3273 cm−1 in the spectra of PEI is from the –N–H– stretching of the molecule, and the peak at 1763 cm−1 in the PLGA spectrum is characteristic of the carbonyl group (–C=O– stretching) present in it. Most importantly, the conjugation of PEI with PLGA (PPN) is confirmed by peaks at 1627 cm−1, of amide I, and 1579 cm−1, of amide II, present in PPN. The glass transition temperature (Tg) of free PLGA and PEI was compared to PPN to observe effective conjugation of PEI to PLGA nanoparticles by DSC analysis. Free PLGA nanoparticles showed a DSC thermogram of 51.15℃, and PEI showed 78.28℃. Conjugation of PLGA to PEI resulted in a shift of Tg to 117.75℃ and an additional Tg of 215.49℃ (Figure 2B). 1H NMR spectroscopy was also used to describe the conjugation of PEI with PLGA (Figure 2C). Peaks at 1.74 ppm correspond to –OH proton, at 1.28 ppm to –CH3, at 3.96 ppm to CH proton and at 3.8 ppm to –CH2 protons of the PLGA unit. While peaks at 2.5 ppm and 2.51 ppm represent –CH2 protons of –NCH2CH2NH2, peaks at 3.17 ppm and 3.66 ppm represent CH2 protons connected with amide and peak at 3.35 ppm represents amino protons of PEI unit. Most importantly, the peak at 7.83 ppm symbolises –RCONH amide proton (Figure 2D), thus confirming the formation of PLGA-PEI conjugate.21
Characterization of Nanoparticles
Particle size and PDI of the particles were measured using a dynamic light scattering system (DLS). Pep5-PLGA particles showed 275.9 ± 12.19 nm, and when PEI and TKD are conjugated, the particle size increased to 420.8 ± 14.72 nm (Figure 3A and B) with a high degree of homogeneity. The increase in the size of nanoparticles indicates that PEI and TKD are conjugated with PLGA effectively. Particle size and morphology analysis of these particles using TEM confirm DLS findings (Figure 3C and D). PLGA nanoparticles and Pep5-TPPN were then subjected to SEM imaging to examine the surface morphology (Figure 3E and F). Spherical PPNs were found in the SEM micrograph. The size distributions confirm the measurements by DLS and TEM. This small size of the particles may aid in improving the uptake of nanoparticles to tumor microenvironment by enhanced permeability and retention effect.30 Surface charges of Pep5-TPPN were analysed before and after PEI and TKD conjugation, and obtained values are −11.6 ± 2.16 mV and +20.01 ± 2.97 mV, respectively. The negative charge of Pep5-PLGA nanoparticles can be attributed to the free carboxyl group of PLGA, and when modified with PEI and TKD, its surface charge is shifted to a positive value. This change in surface charge is because of free amino groups present in PEI (Figure 3G and H).
Pep5 was loaded on PLGA by solvent evaporation method followed by PEI and TKD conjugation using EDC-NHS chemistry. Unloaded Pep5, unreacted PEI and TKD were removed by washing. Overall procedures gave a quite good yield of 67.77 ± 4.29%. Pep5-TPPN were then subjected to peptide loading and encapsulation efficiency calculation. UV spectrophotometric analysis showed that the peptide loading and encapsulation efficiency of Pep5-TPPN were 4.58 ± 0.4% and 77.31 ± 2.69, respectively. High encapsulation efficiency observed results from the hydrophobicity of Pep5.31 Moreover, Pep5 release profile from Pep5-TPPN was also evaluated. At 96 hours, Pep5-TPPN released 81.97 ± 2.83% Pep5 at pH 7.4 in contrast to 96.03 ± 2.92% at pH 6.0 (Figure 3I). This increased Pep5 release at pH 6.0 is attributed to the hydrolysis of biopolymeric nanoparticles in the acidic environment. PVA stabilised PLGA nanoparticles were reported to show similar release kinetics.32
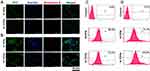
Cell Uptake Study
The cellular internalisation of R-PPN and R-TPPN in MCF7 cells and MDA-MB-231 (stably expressing plasma membrane-targeted EGFP) cells were visualized via CLSM (Figure 4A and B) flow cytometry (Figure 4C and D). Both analyses showed that R-PPN and R-TPPN were internalized into MCF7 and MDA-MB-231 cells within four incubation hours. Over the course of incubation, 70.3 ± 1.89% R-TPPN was accumulated inside MCF7 cells and 84.8 ± 2.45% in MDA-MB-231 cells. Further internalisation was higher with TKD conjugated nanoparticles compared to R-PPN in both cells (68.5 ± 3.19% and 73.1 ± 2.82% in MCF7 and MDAMB231 cells, respectively). So, TKD conjugated nanoparticles were readily internalized into cancer cells by receptor mediated endocytosis. This finding suggests that the cellular uptake capability is enhanced by TKD, which supports the targeting ability of TKD peptide. This increase in Rhodamine-B internalisation by tumour cells when it is delivered actively using tumor targeting peptides is in accordance with previous tumor targeted nano complexes.33,34 The findings further prove the effective conjugation of TKD with PPN.
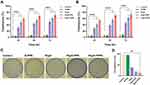
Cytotoxicity Analysis by MTT
The preliminary insight into the anticancer potential of Pep5-PPNs was provided by estimating the cytotoxicity induced by the formulations in MCF7 and MDA-MB-231 cells. To compare the cytotoxic efficacy, both cells were exposed to Pep5, Pep5-PPN, and Pep5-TPPN (at peptide concentration equal to 100 μM) for 24, 48, and 72 h. All the tested formulations showed a time-dependent increase in cytotoxicity on both the cell lines in a time-dependent manner (Figure 5A and B). Moreover, the conjugation of TKD significantly increased the anticancer potential of Pep5-PPN, which matches with the enhanced cytotoxicity induced by targeting peptide conjugated delivery systems.25,29,35 Cells internalised Pep5-PPN and Pe5-TPPN by nonspecific endocytosis and receptor mediated endocytosis, respectively. This could be a plausible explanation for this enhanced cytotoxicity, as per previous reports.36 The high amount of cytotoxicity induced by TKD decorated particles further agrees with the observations made by CLSM analysis. In order to be a successful and safe carrier for therapeutics, it should not impart any cytotoxicity.37 Here, the viability of both cell lines was not affected by B-PPN, which points to the potential of PPN as a successful carrier with high biocompatibility.
Clonogenic Assay
The potential of Pep5-NPs to downregulate the colony forming ability of MCF7 cells was examined by clonogenic assay. Compared to untreated control and Pep5 alone treated cells, the clonogenicity potential of Pep5-NPs treated cells was significantly reduced. The size and number of colonies also significantly declined following treatment with Pep5-NPs (Figure 5C). Pep5-TPPN treated cells showed a decrease in clonogenicity index of 3.6-fold and 0.5-fold with respect to Pep5 and Pep5-PPN treated cells (Figure 5D). This observation lies with previous reports, where successful treatment strategies could efficiently decrease the clonogenicity of the tumor cells.38–40 These findings suggest that Pep5-TPPN can be an effective treatment modality in inhibiting tumor growth.
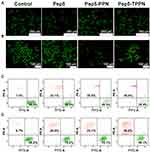
Live Dead Assay
Live dead assay was performed further to confirm the antitumor potential of Pep5-NPs. The assay is based on intracellular esterase activity and plasma membrane integrity.41 Esterase activity in the live cell converts nonfluorescent cell-permeable calcein to the intensely fluorescent calcein. This gives green fluorescence to the cell as they remain within the live cell. Ethidium homodimer (EthD) enters the cells with damaged membranes and binds to nucleic acids, producing a red fluorescence in dead cells. Cell viability, when assessed by live dead assay, all Pep5 formulations induced potent cytotoxicity in both breast cancer cell lines within 48 h of treatment compared to untreated control. This is evidenced by a reduction in the number of green fluorescent cell bodies (living) and an increase in the number of red fluorescent nuclei (dead) (Figure 6A and B). These observations were further validated by flow cytometry analysis, and the results were consistent with the earlier findings (Figure 6C and D). In both experiments, the population of dead cells was higher in Pep5-TPPN treated cells than Pep5 alone and Pep5-PPN treated cells, which substantiates the potential of Pep5-TPPN for successful cancer targeting. In addition, the cytotoxic potency of Pep5 is boosted when entrapped in PPNs, which further increased on conjugation with TKD on the surface of Pep5-TPPN. Similar findings are reported where Pep5 is hybridised with different cell penetrating peptides and endosomal escape peptides for their stimulated cytotoxicity.42
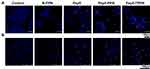
Lysosomal Integrity Assessment Using Acridine Orange Staining
Pep5-NPs were tested for their ability to impair the integrity of the lysosomal membrane by acridine orange staining of the treated cells. Acridine orange is trapped inside the lysosome and displays red fluorescence against a green background as it is protonated in the lysosomal acidic pH. On the other hand, impaired lysosomes have an alkaline pH, which results in a shift in fluorescence from red to green, just like other cellular organelles.24 There is a significant reduction of red fluorescence in Pep5, Pep5-PPN, and Pep5-TPPN treated MCF7 and MDA-MB-231 cells compared to untreated control cells (Figure 7A and B). These observations revealed nanoparticle-mediated destabilisation of the lysosomal membrane in all the treated cells, with Pep5-PPN and Pep5-TPPN treated cells showing the most substantial damage. Inside lysosome, PEI molecules conjugated to the PLGA nanoparticles can induce influx of protons and their counter ions, thereby causing the rupture of lysosomes because of osmotic swelling.43 The lysosomal dysfunction visualised is related to cell death,44 which further correlates with the increased cytotoxicity of Pep5-PPN and Pep5-TPPN.
Assessment of Cytoskeletal Integrity Using Rhodamine Phalloidin Staining
Perturbations of the cytoskeletal system induced by Pep5 and Pep5-NPs were investigated by rhodamine phalloidin staining and a confocal laser scanning microscope. Upon exposure to Pep5, Pep5-PPN and Pep5-TPPN, both MCF7 and MDA-MB-231 cells exhibited a remarkable change in morphology compared to the control cells. This indicates Pep5 and Pep5 loaded nanoformulations significantly affected the integrity of actin filaments. Moreover, stained cells displayed more bundled and less diffused actin filaments in all the treatment groups compared to that in control (Figure 7C and D). These findings are in corroboration with previous research, where Pep5 was shown to induce cytoskeletal damage in MDA-MB-231 cells when fused with a cell penetrating peptide. Other anticancer peptides were also reported to change the microfilament organisation.26 The loss of integrity of actin filament could be attributed to the binding of Pep5 to Plectin, which hampers their ability to organize the cytoskeleton by interfering with the protein–protein interaction.6,45
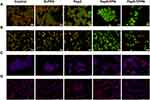
Chromosome Condensation Assay
Since it is evident that Pep5-NPs exert significant cytotoxicity in the tested cells, they were tested for their ability to induce apoptosis. As condensation and fragmentation of chromosomes accompany apoptosis,13 chromatin condensation in MCF7 and MDA-MB-231 cells was investigated by DAPI staining after exposing the cells to Pep5, Pep5-PPN and Pep5-TPPN (Figure 8A and B). After 48 hours of incubation, all the treated groups displayed cells with nuclear condensation and fragmentation. In contrast, untreated cells show no evidence of chromosomal condensation or fragmentation. Nuclear condensation was significantly higher in Pep5-TPPN exposed cells compared to Pep5 and Pep5-PPN exposed cells. The presence of condensed and fragmented chromosomes visualised by DAPI staining suggests the triggering of apoptosis in the cells treated with Pep5 formulations.
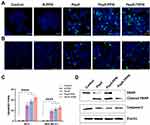
Analysis of Apoptosis by Immunofluorescence for Caspase-3
In order to ascertain the mechanism of cell death triggered by Pep5 and Pep5-NPs in breast cancer cells, immunocytochemistry analysis was conducted for cleaved Caspase-3 expression. Discharge of cytochrome C from mitochondria initiates the caspase cascade to induce apoptotic cell death.46 Cleavage of Caspase-3, an effector Caspase, is a crucial step in executing apoptosis. Effector Caspase brings about the changes during apoptosis, including fragmentation of DNA, dismantling of cytoskeletal proteins and cleavage of PARP.47 Pep5 mediated activation of cleaved Caspase-3 was described earlier.4 Expression of cleaved Caspase-3 is manifested in MCF-7 and MDA-MB-231 cells when treated with Pep5, Pep5-PPN and Pep5-TPPN, as evidenced by the green fluorescence (Figure 9A and B). In contrast, untreated control cells remain with no signs of apoptosis. The degree of Caspase-3 cleavage after Pep5-NPS treatment is depicted as the apoptotic index (Figure 9C).48 Pep5-TPPN treated cells showed a higher apoptotic index than Pep5 alone and Pep5-PPN treated cells. These observations suggest that Pep5 induced cell death by apoptosis is stimulated when Pep5 is entrapped in PPNs, which is further potentiated when Pep5-PPN is conjugated to TKD. In addition, these observations are in line with the previous findings.
Western Blotting
The mechanistic insight of cell death induced by Pep5 and Pep5-NPs was provided by Western blotting. Western blot analysis showed Cleavage of PARP and Caspase-3, the hallmark of apoptosis (Figure 9D).49,50 During apoptosis, Caspase-3 activation leads to the cleavage of PARP.51,52 The decreased expression of Caspase-3 in all treated cells compared to untreated control cells demonstrated the cleavage of Caspase-3, as is visualised by cleaved Caspase-3 immunofluorescence. Furthermore, an increased expression of cleaved PARP was visualised in cells subjected to treatment with Pep5-PPN and Pep5-TPPN. Interestingly, PARP was completely cleaved in Pep5-TPPN exposed cells, indicated by the absence of intact PARP in these cells. This finding prompted the fact that TKD conjugation had significantly increased the therapeutic potential of developed Pep5-NPs by the specific targeting of Pep5 to memHSP70 expressed tumor cells. The cleavage of Caspase-3 and PARP following the treatment with Pep5 and Pep5-NPs could be attributed to the activation of the intrinsic or extrinsic pathway of apoptosis, as Caspase-3 cleavage is a common step of these pathways.47,53,54
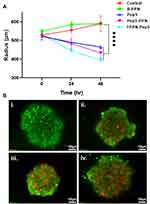
Ex vivo Anti-Cancer Therapeutic Efficacy
Further, to explore the therapeutic potential of Pep5-NPs, 3D tumor models were employed. Tumor spheroids are a perfect ex vivo model for the analysis of new cancer therapeutics as they bear a strong resemblance with the heterogeneity and the conditions of tumor microenvironment in vivo.22,55,56 3D MCF7 spheroids of uniform diameter were developed and treated with Pep5, and Pep5-NPs for 48 hours. This treatment caused significant damage to the spheroids, indicated by the reduction in the size of spheroid over the treatment groups from 0 to 48th hours (Figure 10A). Although the cancer spheroids exposed to Pep5 alone reduced the tumor size, the reduction was more prominent in Pep5-PPN and Pep5-TPPN treated spheroids, being Pep5-TPPN had more impact as with the above results. This could be due to poor penetration of Pep5 into the spheroids. The penetration might have increased upon entrapment of Pep5 in PPN and TPPN. The core of the tumor is believed to trigger angiogenesis, resistance and metastasis, making it a crucial target for successful cancer treatment.57 Furthermore, the efficacy of Pep5-TPPN in targeting the spheroid core was examined using a live dead assay. The Pep5-TPPN treated group has a deteriorated spheroid core, as seen by the elevated red fluorescence with respect to Pep5/Pep5-PPN treated group (Figure 10B). This cell death induced in the spheroids was significantly higher than that induced by other anticancer peptide-based systems.26 These observations were finally substantiated cell viability data observed with MTT assay and live/dead assay. These findings highlight the potential of Pep5-TPPN as an effective therapeutic approach for breast cancer treatment.
Conclusions
Pep5 encapsulated PLGA-PEI nanoparticles were fabricated for safe delivery of Pep5 to ER+ve and TNBC cells. These particles were further coupled to TKD on their surface for targeted delivery of Pep5, yielding a dual peptide functionalised multi-layered nanodrug delivery system. These actively targeted nanoparticles showed enhanced cellular uptake leading to increased Pep5 induced cell death in breast tumor cells. Cytoskeletal disruption and cleavage of both Caspase-3 and PARP demonstrated the sustained mechanism of action of Pep5. Ex vivo studies in tumor spheroids showed significant inhibition of tumor growth when Pep5 was delivered actively. Therefore, Pep5 claims an attractive position in the development of new cancer therapeutics. Moreover, Pep5-TPPN expand the scope of targeted delivery of antitumor peptides for tumor therapy.
Acknowledgments
We wish to thank University Grants Commission (UGC), New Delhi for providing Junior Research Fellowship to Akhil K Mohan and Department of Biotechnology (DBT) for funding.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
2. Miller E, Lee HJ, Lulla A, Hernandez L, Gokare P, Lim B. Current treatment of early breast cancer: adjuvant and neoadjuvant therapy. F1000 Res. 2014;3:1.
3. Xiao YF, Jie MM, Li BS, et al. Peptide-based treatment: a promising cancer therapy. J Immunol Res. 2015;2015:1.
4. de Araujo CB, Russo LC, Castro LM, et al. A novel intracellular peptide derived from g1/s cyclin d2 induces cell death. J Biol Chem. 2014;289(24):16711–16726.
5. Chen BB, Glasser JR, Coon TA, et al. F-box protein FBXL2 targets cyclin D2 for ubiquitination and degradation to inhibit leukemic cell proliferation. Blood J Am Soc Hematol. 2012;119(13):3132–3141.
6. Russo LC, Araujo CB, Iwai LK, Ferro ES, Forti FL. A Cyclin D2-derived peptide acts on specific cell cycle phases by activating ERK1/2 to cause the death of breast cancer cells. J Proteomics. 2017;151:24–32.
7. Yang HY, Meng J, Jang MS, et al. CD44-targeted and enzyme-responsive photo-cross-linked nanogels with enhanced stability for in vivo protein delivery. Biomacromolecules. 2021;22(8):3590–3600.
8. Patra JK, Das G, Fraceto LF, et al. Nano based drug delivery systems: recent developments and future prospects. J Nanobiotechnology. 2018;16(1):1–33.
9. Makadia HK, Siegel SJ. Poly lactic-co-glycolic acid (PLGA) as biodegradable controlled drug delivery carrier. Polymers. 2011;3(3):1377–1397.
10. Maiolino S, Russo A, Pagliara V, et al. Biodegradable nanoparticles sequentially decorated with Polyethyleneimine and Hyaluronan for the targeted delivery of docetaxel to airway cancer cells. J Nanobiotechnology. 2015;13(1):1–13. doi:10.1186/s12951-015-0088-2
11. Fasehee H, Dinarvand R, Ghavamzadeh A, et al. Delivery of disulfiram into breast cancer cells using folate-receptor-targeted PLGA-PEG nanoparticles: in vitro and in vivo investigations. J Nanobiotechnology. 2016;14(1):1–18. doi:10.1186/s12951-016-0183-z
12. Zhang L, Jin D, Stenzel MH. Polymer-functionalized upconversion nanoparticles for light/imaging-guided drug delivery. Biomacromolecules. 2021;22(8):3168–3201. doi:10.1021/acs.biomac.1c00669
13. Geethakumari D, Sathyabhama AB, Sathyan KR, Mohandas D, Somasekharan JV, Puthiyedathu ST. Folate functionalized chitosan nanoparticles as targeted delivery systems for improved anticancer efficiency of cytarabine in MCF-7 human breast cancer cell lines. Int J Biol Macromol. 2022;199:150–161. doi:10.1016/j.ijbiomac.2021.12.070
14. Sun B, Zhang S, Zhang D, et al. Identification of metastasis-related proteins and their clinical relevance to triple-negative human breast cancer. Clin Cancer Res. 2008;14(21):7050–7059. doi:10.1158/1078-0432.CCR-08-0520
15. Stangl S, Gehrmann M, Riegger J, et al. Targeting membrane heat-shock protein 70 (Hsp70) on tumors by cmHsp70.1 antibody. Proc Natl Acad Sci. 2011;108(2):733–738. doi:10.1073/pnas.1016065108
16. Gehrmann M, Stangl S, Foulds GA, et al. Tumor imaging and targeting potential of an Hsp70-derived 14-mer peptide. PLoS One. 2014;9(8):e105344. doi:10.1371/journal.pone.0105344
17. Elmallah MIY, Cordonnier M, Vautrot V, Chanteloup G, Garrido C, Gobbo J. Membrane-anchored heat-shock protein 70 (Hsp70) in cancer. Cancer Lett. 2020;469:134–141. doi:10.1016/j.canlet.2019.10.037
18. Gastpar R, Gross C, Rossbacher L, Ellwart J, Riegger J, Multhoff G. The cell surface-localized heat shock protein 70 epitope TKD induces migration and cytolytic activity selectively in human NK cells. J Immunol. 2004;172(2):972–980. doi:10.4049/jimmunol.172.2.972
19. Meng Y, Wang S, Li C, et al. TKD peptide as a ligand targeting drug delivery systems to memHsp70-positive breast cancer. Int J Pharm. 2016;498(1–2):40–48.
20. Gu P, Wusiman A, Wang S, et al. Polyethylenimine-coated PLGA nanoparticles-encapsulated Angelica sinensis polysaccharide as an adjuvant to enhance immune responses. Carbohydr Polym. 2019;223:115128.
21. Lü JM, Liang Z, Wang X, Gu J, Yao Q, Chen C. New polymer of lactic-co-glycolic acid-modified polyethylenimine for nucleic acid delivery. Nanomedicine. 2016;11(15):1971–1991.
22. Gupta PK, Tripathi SK, Pappuru S, et al. Metal-free semi-aromatic polyester as nanodrug carrier: a novel tumor targeting drug delivery vehicle for potential clinical application. Mater Sci Eng C. 2020;107:110285.
23. Chakravarthi SS, Robinson DH. Enhanced cellular association of paclitaxel delivered in chitosan-PLGA particles. Int J Pharm. 2011;409(1–2):111–120.
24. Athira SS, Biby TE, Mohanan PV. Effect of polymer functionalized fullerene soot on C6 glial cells. Eur Polym J. 2020;127:109572.
25. Jung HK, Kim S, Park RW, Park JY, Kim IS, Lee B. Bladder tumor-targeted delivery of pro-apoptotic peptide for cancer therapy. J Control Release. 2016;235:259–267.
26. Hadianamrei R, Tomeh MA, Brown S, Wang J, Zhao X. Rationally designed short cationic α-helical peptides with selective anticancer activity. J Colloid Interface Sci. 2022;607:488–501.
27. Hilchie AL, Sharon AJ, Haney EF, et al. Mastoparan is a membranolytic anti-cancer peptide that works synergistically with gemcitabine in a mouse model of mammary carcinoma. Biochim Biophys Acta. 2016;1858(12):3195–3204.
28. Shahin M, Ahmed S, Kaur K, Lavasanifar A. Decoration of polymeric micelles with cancer-specific peptide ligands for active targeting of paclitaxel. Biomaterials. 2011;32(22):5123–5133.
29. Mori T, Hazekawa M, Yoshida M, Nishinakagawa T, Uchida T, Ishibashi D. Enhancing the anticancer efficacy of a LL-37 peptide fragment analog using peptide-linked PLGA conjugate micelles in tumor cells. Int J Pharm. 2021;606:120891.
30. Lee C, Kang S. Development of HER2-targeting-ligand-modified albumin nanoparticles based on the SpyTag/SpyCatcher system for photothermal therapy. Biomacromolecules. 2021;22(6):2649–2658.
31. Zhang Q, Li D, Guan S, et al. Tumor-targeted delivery of honokiol via polysialic acid modified zein nanoparticles prevents breast cancer progression and metastasis. Int J Biol Macromol. 2022;203:280–291.
32. Bomb RSP. Dual drug delivery of curcumin and niclosamide using PLGA nanoparticles for improved therapeutic effect on breast cancer cells. J Polym Res. 2020;27(5):1–13.
33. Bang JH, Ryu YC, Kim KA, Hwang BH. Targeted delivery of self-assembled nanocomplex between fusion peptides and siRNAs for breast cancer treatment. Biochem Eng J. 2022;186:108564.
34. Hazeri Y, Samie A, Ramezani M, et al. Dual-targeted delivery of doxorubicin by mesoporous silica nanoparticle coated with AS1411 aptamer and RGDK-R peptide to breast cancer in vitro and in vivo. J Drug Deliv Sci Technol. 2022;71:103285.
35. Nabi PN, Vahidfar N, Tohidkia MR, Hamidi AA, Omidi Y, Aghanejad A. Mucin-1 conjugated polyamidoamine-based nanoparticles for image-guided delivery of gefitinib to breast cancer. Int J Biol Macromol. 2021;174:185–197.
36. Chen G, Jaskula–Sztul R, Harrison A, et al. KE108-conjugated unimolecular micelles loaded with a novel HDAC inhibitor thailandepsin-A for targeted neuroendocrine cancer therapy. Biomaterials. 2016;97:22–33.
37. Tian F, Dahmani FZ, Qiao J, et al. A targeted nanoplatform co-delivering chemotherapeutic and antiangiogenic drugs as a tool to reverse multidrug resistance in breast cancer. Acta Biomater. 2018;75:398–412.
38. Bhavsar DB, Patel V, Sawant KK. Design and characterization of dual responsive mesoporous silica nanoparticles for breast cancer targeted therapy. Eur J Pharm Sci. 2020;152:105428.
39. Faheem MM, Bhagat M, Sharma P, Anand R. Induction of p53 mediated mitochondrial apoptosis and cell cycle arrest in human breast cancer cells by plant mediated synthesis of silver nanoparticles from Bergenia ligulata (Whole plant). Int J Pharm. 2022;2022:121710.
40. Pillai JJ, Thulasidasan AKT, Anto RJ, Devika NC, Ashwanikumar N, Kumar GSV. Curcumin entrapped folic acid conjugated PLGA–PEG nanoparticles exhibit enhanced anticancer activity by site specific delivery. RSC Adv. 2015;5(32):25518–25524.
41. Pfeffer BA, Fliesler SJ. Streamlined duplex live-dead microplate assay for cultured cells. Exp Eye Res. 2017;161:17–29.
42. Wang T, Tian L, Cheng Q, et al. Pep5-based antitumor peptides containing multifunctional fragments with enhanced activity and synergistic effect. Eur J Med Chem. 2022;2022:114320.
43. Meng F, Kwon S, Wang J, Yeo Y. Immunoactive drug carriers in cancer therapy. In: Biomaterials for Cancer Therapeutics. Elsevier; 2020:53–94.
44. Boya P. Lysosomal function and dysfunction: mechanism and disease. Antioxid Redox Signal. 2012;17(5):766–774.
45. Kazerounian S, Uitto J, Aho S. Unique role for the periplakin tail in intermediate filament association: specific binding to keratin 8 and vimentin. Exp Dermatol. 2002;11(5):428–438.
46. Redza-Dutordoir M, Averill-Bates DA. Activation of apoptosis signalling pathways by reactive oxygen species. Biochim Biophys Acta Mol Cell Res. 2016;1863(12):2977–2992.
47. Mata R, Nakkala JR, Sadras SR. Polyphenol stabilized colloidal gold nanoparticles from Abutilon indicum leaf extract induce apoptosis in HT-29 colon cancer cells. Colloids Surf B. 2016;143:499–510.
48. Wanjale MV, Jaikumar VS, Sivakumar KC, Paul RA, James J, Kumar GSV. Supramolecular hydrogel based post-surgical implant system for hydrophobic drug delivery against glioma recurrence. Int J Nanomedicine. 2022;17:2203.
49. Chowdhury P, Nagesh PKB, Hatami E, et al. Tannic acid-inspired paclitaxel nanoparticles for enhanced anticancer effects in breast cancer cells. J Colloid Interface Sci. 2019;535:133–148.
50. Wang X, Yang L, Zhang H, et al. Fluorescent magnetic PEI-PLGA nanoparticles loaded with paclitaxel for concurrent cell imaging, enhanced apoptosis and autophagy in human brain cancer. Colloids Surf B. 2018;172:708–717.
51. To TL, Shu X. Major methods and technologies for assessing cell death. In: Mechanisms of Cell Death and Opportunities for Therapeutic Development. Elsevier; 2022:93–118.
52. Charriaut-Marlangue C, Renolleau S. Models for apotosis: from newborn to adult; 2006.
53. Ketelut-Carneiro N, Fitzgerald KA. Apoptosis, pyroptosis, and necroptosis—Oh my! The many ways a cell can die. J Mol Biol. 2022;434(4):167378.
54. Ghosh S, Kundu M, Dutta S, et al. Enhancement of anti-neoplastic effects of cuminaldehyde against breast cancer via mesoporous silica nanoparticle based targeted drug delivery system. Life Sci. 2022;2022:120525.
55. Lazzari G, Couvreur P, Mura S. Multicellular tumor spheroids: a relevant 3D model for the in vitro preclinical investigation of polymer nanomedicines. Polym Chem. 2017;8:4947–4969.
56. Van Zundert I, Fortuni B, Rocha S. From 2D to 3D cancer cell models—The enigmas of drug delivery research. Nanomaterials. 2020;10:2236.
57. Ps R, Alvi SB, Begum N, Veeresh B, Rengan AK. Self-assembled fluorosome–polydopamine complex for efficient tumor targeting and commingled photodynamic/photothermal therapy of triple-negative breast cancer. Biomacromolecules. 2021;22(9):3926–3940.
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.