Back to Journals » Biologics: Targets and Therapy » Volume 16
Phage Therapy: A Different Approach to Fight Bacterial Infections
Authors Hibstu Z , Belew H , Akelew Y, Mengist HM
Received 5 July 2022
Accepted for publication 22 September 2022
Published 6 October 2022 Volume 2022:16 Pages 173—186
DOI https://doi.org/10.2147/BTT.S381237
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Doris Benbrook
Zigale Hibstu, Habtamu Belew, Yibeltal Akelew, Hylemariam Mihiretie Mengist
Department of Medical Laboratory Science, College of Health Sciences, Debre Markos University, Debre Markos, Ethiopia
Correspondence: Zigale Hibstu, Email [email protected]
Abstract: Phage therapy is one of the alternatives to treat infections caused by both antibiotic-sensitive and antibiotic-resistant bacteria, with no or low toxicity to patients. It was started a century ago, although rapidly growing bacterial antimicrobial resistance, resulting in high levels of morbidity, mortality, and financial cost, has initiated the revival of phage therapy. It involves the use of live lytic, bioengineered, phage-encoded biological products, in combination with chemical antibiotics to treat bacterial infections. Importantly, phages will be removed from the body within seven days of clearing an infection. They target specific bacterial strains and cause minimal disruption to the microbial balance in humans. Phages for medication must be screened for the absence of resistant genes, virulent genes, cytotoxicity, and their interaction with the host tissue and organs. Since they are immunogenic, applying a high phage titer for therapy exposes them and activates the host immune system. To date, no serious side effects have been reported with human phage therapy. In this review, we describe phage–phagocyte interaction, bacterial resistance to phages, how phages conquer bacterial resistance, the role of genetic engineering and other technologies in phage therapy, and the therapeutic application of modified phages and phage-encoded products. We also highlight the comparison of antibiotics and lytic phage therapy, the pros and cons of phage therapy, determinants of human phage therapy trials, phage quality and safety requirements, phage storage and handling, and current challenges in phage therapy.
Keywords: lysogenization, lytic phage, modified phages, resistance to phages, CRISPR, immunity, conquering CRISPR, phage-encoded products
Introduction
Phages are obligate intracellular viruses that infect and kill bacteria. They exist everywhere that bacteria live and there are about 1029–1030 phages in the biosphere. They have high durability in natural systems and the inherent potential to reproduce rapidly in their appropriate host.1,2 Phages are made up of proteins or proteolipid capsids containing fragments of deoxynucleic acid (DNA) or ribonucleic acid (RNA) (Figure 1). Their genome size ranges from a few thousand to 498 kbs.3 Phages have no machinery to generate energy or ribosomes to make proteins, even though they carry the genetic information needed to replicate in the right host cell.
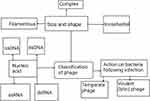 |
Figure 1 Classification of phages. |
Phages are generally specific and their specificity is determined by phage–host receptor surface, genetic and host physical defense mechanisms, the nature of the phage(s), and their co-evolution. Phage lytic enzymes (endolysins) have broader specificity at the genus and/or species level. However, their specificity varies from infecting many bacteria to infecting a single strain. Limitations in sensitivity to a single phage therapy during polymicrobial infection are solved by applying phage cocktails.4
Phage therapy is a way of delivering virulent phages to a clinically ill patient to rapidly kill pathogenic bacteria.5 It involves the use of lytic phages, bioengineered phages, and purified lytic proteins of phages to infect and lyse bacteria at the site of infection. Phages and their lytic proteins can be used specifically to treat multidrug-resistant (MDR) bacteria, either alone or supplemented with antibiotics. Therapeutic approaches using phages are rapidly increasing, although there is still no adequate knowledge on phage–phage, phage–bacteria, or phage–human interactions, mainly because of safety and efficacy concerns.6 Novel concepts in phage therapy involve direct treatment of bacterial infections, phage-mediated prevention of bacterial infection, and the exploration of phage diversity in environmental and human ecological niches (Figure 2).5 Currently, human phage therapy trials are being undertaken, although therapeutic use of phages is limited to Georgia, Poland, and Russia.7 Phage therapy is a promising approach to fighting bacterial infections, as phages have unique bacteria-killing mechanisms and life cycles; either lytic or lysogenic growth cycles (Figure 3). Only lytic phages are used for therapeutic purposes. They inhibit the emergence of resistant bacteria by killing the bacteria that they infect and are preferable to antibiotics as they cause less damage to the general microbiome.8 Lytic phage therapy involves the replication of phages in phage-infected bacteria; the phages disrupt bacterial metabolism and kill the bacteria.
 |
Figure 2 Novel concepts of phage therapy. |
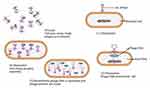 |
Figure 3 Phage lytic life cycle. Notes: Adapted from Adesanya O, Oduselu T, Akin-Ajani O, Adewumi OM, Ademowo OG. An exegesis of bacteriophage therapy: An emerging player in the fight against anti-microbial resistance. AIMS Microbiol. 2020;6(3):204–230 (https://creativecommons.org/licenses/by/4.0/).104 |
The ability of most lytic phages to encode the enzymes, holins and endolysins, that degrade bacterial structures (cell membrane and cell wall) make them a potential new weapon in the fight against bacterial infections. This property makes them efficacious against both antibiotic-sensitive and antibiotic-resistant bacteria. However, some lytic phages use only endolysins. Indeed, holins degrade the bacterial cytoplasm, allowing endolysins to access bacterial glycoproteins. Holins control the exact point in time for endolysins to access the bacterial murein and synchronize the holin–endolysin system to the late stage of viral replication. The synergetic holin–endolysin system causes cell lysis and the release of mature lytic phage progeny.9 Thereby, 50–200 mature phage progenies will be released from the lysis of a bacterium.11,45
Temperate phages are not used for therapeutic purposes because they integrate their genome into the host chromosome or sometimes maintain it as a plasmid to be transmitted to daughter cells during cell division or horizontally across the bacterial community. They may undergo a typical lytic cycle or lysogenization. Temperate phages enter the lytic life cycle when host conditions are weakened, maybe due to a scarcity of nutrients; then, prophages become active. At this stage, they promote the reproductive cycle, resulting in lysis of the bacterial cell. In the lysogenic life cycle, the virus continues to replicate as the bacterium continues to reproduce, and is found in all bacterial offspring. Example, phage lambda of E. coli is a common phage that has both a lysogenic cycle and a lytic cycle.10 Surprisingly, temperate phages may increase the pathogenicity of the host bacteria because bacterial virulent genes are identified from their genome.11
Phage cocktails are used for phage therapy owing to rapidly emerging bacterial resistance, because many types of phages infect the same species or strain of bacterium. Thus, phage cocktails could be used to target different structural sites and metabolic activities of a bacterium. It may be argued that only a single specific phage should be used against a pathogen to prevent the emergence of resistant bacteria, since the extensive use of phages may promote resistance to phage cocktails.12 Some of the challenges in applying phage cocktail therapy are the inability to predict the effect of mass use of phages, the very high cost of phage treatment, the issue of efficacy, and the high specificity of phages.13 The highly increased emergence and spread of resistant pathogens and the lack of new drug production have directed many institutions and commercial companies to become engaged in phage therapy.14 Antibiotic-resistant opportunistic pathogens are a threat, especially for immune-compromised and immune-incompetent patients in healthcare settings. These are serious problems in medicine, to which phage therapy may provide a solution.15
Although the use of phage therapy against bacterial infections is very promising, with plenty of advantages, many advances will be needed to implement phage therapy on a large enough scale for therapeutic purposes, owing to emerging issues on the safety, quality, and stability of phages, and the lack of sufficient evidence for their use in human medication.16
Phage–Phagocyte Interaction
A phage selected for phage therapy should be resistant to phagosomal degradation, to avoid or delay the induction of the phage’s specific adaptive immunity response and extend the survival of the phage in immunocompetent individuals.17,18 Therapeutic phages are naturally immunogenic, so they stimulate complex interactions between innate and adaptive immune cells that may affect the phage therapy. Bacterial elimination occurs owing to stimulation of local immune responses as a result of phage and bacterial-derived pathogen-associated molecular patterns (PAMPs). Since phages are immunogenic, they induce phage-specific humoral memory, which can hamper their therapeutic success owing to neutralization.19
The role of phagocytic cells is to recognize and eliminate foreign antigens and to activate the adaptive immune system response whenever necessary. Leukocytes bind to phages in a time-, concentration-, and temperature-dependent manner, and endocytose them (through phagocytosis for particles >500 nM) to remove them. Polymorphonuclear leukocytes and macrophages can degrade phages, and phage degradation is the first step in stimulating antigen presentation and the development of an adaptive immune response.19,20 When phages express proteins that mediate bacteria–phage interaction, they bind together and macrophages become activated. Macrophages phagocytose extracellular bacteria and endocytose the phages along with them. Phagocytosis is stimulated via bacteria and phage-derived PAMPs and continual phagocytosis of phage-infected bacteria occurs. Bacteria and phage-derived PAMPs again co-stimulate macrophage activity.21,22 Antibodies produced against the bacteria opsonize the bacteria and facilitate phagocytosis by macrophages, which promotes bacterial clearance. Phage–antibody complexes bind to Fc receptors on macrophages, which triggers endocytosis and subsequent phage clearance.
In general, for phage therapy to be effective there must be a strong interaction between host-derived ligands and host pattern recognition receptors. Unfortunately, weak pattern recognition receptor activation in immune-deficient individuals affects the individual innate immune response. Thus, clinical studies need to be conducted by enrolling individuals with different immune deficiency states to apply phage therapy.
Phage–Adaptive Immune System Interaction
Phages strongly influence adaptive immunity via their effects on humeral immunity and effector polarization. They modulate the immune response and have a profound effect on the outcome of bacterial infection.23 Individuals exposed to phage therapy or naturally existing phages will clearly develop antibodies because phages are composed of densely packed immunogenic DNA or RNA and a protein coat.22,24
Phages alone are not sufficient to fight bacterial infection. The combined effect of the immune system along with phage therapy is essential to fight bacterial infections. Phages are themselves immunogenic microbes which can activate the human adaptive immune system. Phage-mediated bacterial lysis stimulates the human adaptive immune response, which enhances the efficacy of phage therapy. However, adverse phage treatment may cause toxicity owing to the release of endotoxins as a result of bacterial lysis.19,25
Phage–immune interactions depend on immune recognition through pattern recognition receptors, the immunogenic nature of the phage, and the multiplication rate of the phage. The pattern recognition receptor recruits phagocytes to the site of infection to resolve the infection. It recognizes phage-derived DNA and RNA, resulting in phage-mediated activation of innate immune cells. The commitment of the pattern recognition receptor and the level of immune activation depend on phage type, phage dose, and nucleic acid synthesis activity.26,27 Phages are normally immunogic in nature. So, to evade host immune response, repeated administration of phages is required to clear bacterial infection. Therefore, immunogenicity should be considered before phages are used for therapy.28 Phages were widely administered intravenously decades ago to diagnose and monitor primary and secondary immunodeficiency without reported complications even in patients with prolonged phage survival in their bloodstream. This implies their inherently low toxic effect.46 Fifty healthy volunteers who were not involved in phage therapy or in phage work were evaluated for anti-phage antibody production against phage T4, and they were positive for the naturally occurring phage antibody.22 A remarkable decline in phage activity was observed in 81% of participants seropositive for the phage antibody. In these positive sera, natural IgG antibodies specific to the phage proteins gp23*gp24*Hoc and Soc were identified. These findings show that anti-T4 phage antibodies are frequent in the human population.47 The multiplication rate of phages can be reduced by IgG or IgA. Phages can be removed from the human body by high antibody levels and the Fc receptor-mediated uptake of phage/antibody complexes by macrophages. One of the drawbacks of phage therapy is that phages are immunogenic, so that antibodies are produced against them, which neutralize the phages and hinder infection of a bacterium by phages.19 At present, there is a knowledge gap on whether this regulatory function of anti-phage antibodies can prevent the appearance of resistance to phages and pre-existing immunity to natural phages, affecting phage therapy. More importantly, it is not known which phage-specific factors are responsible for the mechanism of phage clearance.29 To evade adaptive humeral immunity, further work on phage modification, to remove their immunogenicity and retain their lytic effect, is required.
Bacterial Resistance to Phages
Bacteria can develop resistance to phage therapy through spontaneous mutations, acquisition of restriction–modification (RM) systems, adaptive immunity via the clustered regular interspaced short palindromic repeat-associated (CRISPR-Cas) system, plasmids, temperate genes, and mobile genetic islands (that can carry genes coding for resistance to antibiotics).30,31 These mechanisms can be used by a bacterium to target different steps in the phage life cycle, including phage attachment, penetration, replication, and host cell lysis.30 Prominent resistance phenotypes are noticed as a result of distinct resistance mechanisms. There are different prominent resistance phenotypes depending on whether the resistance is partial or complete, the fitness cost associated with resistance, and whether the mutation can be countered by a mutation in the infecting phage.32
Spontaneous bacterial mutation results in the emergence of phage resistance and phage–bacterium co-evolution,33 which may lead to phage resistance by modifying phage-associated receptors on the bacterial surface. Importantly, such alterations may be associated with reduced fitness relative to non-resistant strains.34 When mutation occurs in bacterial lipopolysaccharides, or when the bacterium undergoes impaired growth as a result of mutations in genes involved in essential cell function, phage-resistant bacteria may become less virulent.35
Bacterial RM systems, which are often called primitive immune systems in bacteria, are ubiquitous.36 They are important defense mechanisms against invading phage genomes. They consist of two contrary enzymatic activities: a restriction endonuclease (REase) and a methyltransferase (MTase). The mechanism of bacterial RM systems in defense is through recognition of the methylation status of invading phage genomes. Methylated sequences are recognized as self, while sequences on the invading phage genome lacking methylation are recognized as foreign and are cleaved by the REase. The role of REase is to recognize and cleave non-self-nucleic acid sequences at specific sites, while the role of MTase activity is to ensure identification of self and foreign nucleic acids, by transferring methyl groups to the same specific nucleic acid sequence within the bacterial genome.37
CRISPRs regulate the adaptive immunity of bacteria. Bacteria can develop adaptive immunity against phages by acquiring a unique bit of the phages’ DNA CRISPR-Cas machinery, called spacers, from prior exposure or infection. Bacterial adaptive immunity against phages is different from other defense mechanisms because bacteria are able to recognize prior infections by storing pieces of phage DNA (spacers) in their own DNA to neutralize future infections. Surprisingly, bacteria can not only recognize prior infections by using CRISPR-Cas, but also transfer this experience to future generations.38
Bacterial mobile genetic elements may promote bacterial resistance to phage therapy. They are responsible for the horizontal transfer of phage-resistant bacterial genes among bacteria. For instance, phage resistance-conferring conjugative plasmids can disseminate quickly both within and between bacterial species by expressing mating-pair complexes that are physically in close proximity.39 Regardless of antibiotic selection, antibiotic resistance plasmids survive in abundance in bacterial populations because plasmids cause minimal bacterial fitness cost. In any case, if there is a fitness cost, it will be balanced rapidly by mutation in the phage or bacteria, or in both.40,41 Phages that specifically bind to the mating-pair complex encoded by conjugative, drug resistance-conferring plasmids have the potential to limit the spread of antibiotic resistance-conferring plasmids.
Conquering CRISPR
CRISPR-Cas is a genome editing system found in bacteria that helps a bacterium to defend against phages by inhibiting the integration of phage DNA to a bacterium via CRISPR and endonuclease activity (Cas). Phages undergo point mutation or deletions to escape bacterial adaptive immunity.42 Therefore, CRISPR-Cas (the effector molecule) fails to recognize and cut the specific genomic sequences of phages that had point mutations and/or deletions. This implies that in CRISPR-Cas systems, a single mutation in the protospacer-adjacent motif is enough to avoid targeting.43–45 Unfortunately, some phage mutations may promote CRISPR immunity by enhancing the gaining of many new spacers.48,49 Phages can evade detection by deleting part of or the entire protospacer target. Despite conquering CRISPR, this strategy can have a fitness cost to the phage, depending on the region deleted.50
Phages produce different proteins that inhibit CRISPR-Cas defense, of which anti-CRISPR (Acr) proteins are the most prominent.51 Inhibition mechanisms of CRISPR-Cas defense include blocking target binding by DNA mimicry or steric blocking and the prevention of DNA cleavage by nucleases.52 An emerging way to inhibit CRISPR-Cas activity is by utilizing phages’ subversion of cellular regulatory pathways that bypass CRISPR-Cas activity.53 Phages may possess regulatory protein homologs to bacterial proteins that suppress bacterial defenses, known as bacterial CRISPR-Cas repressors.54 Otherwise, phages can use proteins that bind and inhibit bacterial regulators. Multiple bacteria modify their CRISPR-Cas activity via quorum sensing, but this behavior may be manipulated by phage-encoded proteins. Genomic modifications are another way for phages to escape from CRISPR-Cas attack.55,56 For example, five distinct anti-CRISPR genes are present in P. aeruginosa temperate phages. These genes encode a small protein that can immediately neutralize the immune system of the host by interfering with the formation or action of CRISPR-Cas ribonucleic protein.57
Two general models have been implemented to manage the risk of bacterial resistance to phage therapy. These involve applying phage cocktails and adapting a single phage to each patient.58 Combining many phages in cocktails provides them a wide host range and improves their effectiveness. Synergizing different phages and targeting different receptors on the bacterial surface reduces bacterial resistance to phages. Such an approach has a major benefit for empirical treatment.59 The personalized phage therapy approach utilizes single phages or targeted phage cocktails directly based on the etiologic agent isolated.60 This approach is more flexible with respect to the phage spectrum, and minimizes the emergence of bacterial resistance effectively, but carries a higher cost for treatment.61
In summary, although bacteria have the potential to develop resistance to phage therapy via different mechanisms, phages have several mechanisms by which they can escape bacterial resistance against them. Therefore, upgrading current practices and knowledge on phage interactions with phages, bacteria, and humans, is a promising way to treat bacterial infections in the era of increasing incidence and transmission of MDR bacterial species and strains, where the production of new antibiotics is limited.
Role of Genetic Engineering and Other Genetic Technologies for Phage Therapy
The role of genetic engineering in phage therapy is to produce phages with a broader host range and the recombination of two distinct phages, while the role of synthetic biology is to construct the whole genome of the phages, so that artificial phages that can infect bacteria can be developed. Whole genome sequencing allowed the production of new phage variants with an expanded host range and a few phage strains to cover diversified bacteria.6 Genetic engineering can be used to incorporate bacteriocins, enzybiotics, quorum sensing inhibitors, and biofilm-degrading enzymes into phages. These molecules inhibit bacterial metabolism and other vital bacterial activities. Therefore, when a bacterium is infected by a phage carrying these molecules, it will die. For example, an engineered T7 phage was engineered to encode lactonase. (Lactonase has a broad activity and inhibits quorum sensing molecules in bacteria, required for biofilm formation.)62 Engineered phages improve on conventional methods used to kill bacteria. This implies that phages can be engineered with entirely novel mechanisms to kill bacteria and alter the mode of gene expression of targeted bacteria.63 Recently, a cystic fibrosis patient with a disseminated Mycobacterium abscessus infection was treated by applying engineered bacteriophages for the first time. There are over 1800 mycobacterial phages in the bank, but only one of them effectively killed the clinical isolate of M. abscessus.64
Advances in sequencing technology and synthetic biology have provided new opportunities to modify and use temperate genes to fight the ever-increasing antibiotic resistance.65 The gene editing technology CRISPR is used to target a specific genome sequence for site-specific cleavage. For example, it has been applied to carbapenem-resistant Enterobacteriaceae and enterohemorrhagic E. coli.9
Modified Phages and Their Therapeutic Applications
Bioengineered phages highly minimize the drawbacks of conventional phage therapies owing to their ability to reach and kill the targeted pathogens or reverse the drug resistance of bacteria. Phage-derived biochemical endolysin (phage lysin) is effective against Gram-positive bacteria and it can be used for bacterial treatment instead of viable phages.5,14
Modified phages are phages whose specificity is deliberately altered into non-native forms. Their host recognition specificity is conferred by receptor binding domains found on phages.66 Modified phages are used to target non-native hosts; they are designed to serve as a vehicle into which antimicrobials are incorporated or attached to the surface, to suppress the host SOS DNA repair system.67,68 Modifications enable phages to overcome the narrow host range of phages, and can reduce the potential of bacteria to develop resistance, avoid challenges in phage manufacturing, exclude systemic side effects (especially endotoxin release), and prevent the phage being attacked by the immune system. To avoid the release of endotoxins by Gram-negative bacteria, mainly due to lytic phages or antibiotic treatment, phages can be made lysine deficient. For example, MRSA-infected mice were successfully treated by lysine-deficient phages because the bacterium was killed without lysis. Another means of using modified phages is a targeted gene delivery system to the site of infection using engineered filamentous phages.12 Modified phages can bypass the host immune system, persist within the body, and deliver lethal genes to the bacterial host. Many experiments in animal models revealed that engineered phages are efficient in treating infections. Filamentous phages that do not lyse the host are used as a vehicle to provide lethal genes or substances such as holins, lethal transcription regulators, and addiction toxins to induce apoptosis specifically at the site of infection.69
Phage-Encoded Products
Phages encode proteins that recognize and adhere to sites on the bacterial surface, such as peptidoglycans, pili, flagella, or efflux pumps, and to specific sugar moieties in lipopolysaccharides.70 They encode two types of lysosomes, porin endolysins and phage tail-associated murein lytic enzymes. These enzymes degrade the cell wall of the host. Endolysins, with the help of holin, lyse the bacterial cell wall from the inside and allow phage progenies to be released.71 The ability of endolysins to bind firmly to substrates on the host cell wall minimizes the turnover of endolysins and the requirement of many endolysin molecules to degrade bonds on the cell of the host.72 Phage tail-associated murein lytic enzymes hydrolyze the cell wall after adsorption of the phage to the host cell wall from the outside;73 their activity is limited to Gram-positive bacteria because Gram-negative bacteria have an outer membrane that blocks direct enzyme contact with the peptidoglycan of the host cell wall. Structurally engineered phage lysosomal molecules that have specific binding and fusing ability with other modified lysosomes have shown encouraging results against Gram-negative bacteria.74
Phage-encoded products are used to kill pathogens directly. Their merits over viable phage therapy include their enhanced ability to penetrate and diffuse to the site of action by bypassing sequestration by the spleen, lymph nodes, and other organs. Lysine is an example of a phage product assumed as antibacterial weapons. It is safe and efficient against bacteria and resistance to lysine is less frequent compared to antibiotics. They are successful in animal models against Gram-positive bacteria including S. pneumonia, S. pyogenes, B. anthracis, E. faecium, and S. aureus, but not against Gram-negative bacteria, to date.75
It is unlikely for a bacterium to evolve resistance to lysins, since lysins target sites on the peptidoglycan, which is vital for bacterial cell viability.76,77 In addition, mass preparation and administration of engineered recombinant lytic proteins is much easier than mass preparation and administration of actual phages. Modified products of phages have more potential than natural phages because viable phages have limitations due to their short shelf life, sequestration by the reticuloendothelial system, and potential to induce neutralizing antibodies.78 Applying phage lysin therapy in combination with antibiotics is more effective than the single use of either lysins or antibiotics.79,80
Phage Therapy in Humans
During phage therapy, lytic phages are mainly applied to kill their bacterial hosts with no effect on human cells and no or minimal disturbance to the human microbiota relative to conventional antibiotics. Phage therapy is rapidly being revived, with encouraging effects in life-saving therapeutic use and multiple clinical trials. However, it is meeting obstacles with respect to regulations and policy issues for clinical use and implementation.81
For clinical trials on phage therapy, phages should be sufficiently characterized and there must be selection of the right phage, human, and bacterium, and consideration of the right disease target for phage therapy. In addition, information on phage formulation, dosage, and efficacy is vital for effective therapy. For example, very specific phages are desired for monobacterial disease. However, this may be a limitation during polybacterial infections, unless the phage is provided in combination with antibiotics. This approach is relevant to patient safety because the removal of a single pathogen and the growth of other bacteria may threaten the patient’s life.82 In fact, phages with a broad host range may be more abundant than currently identified phages, although further investigations are required.83 Principally, all precautions, policies, and regulations for phage therapy need standardization, as is the case for antibiotics for human use.
The lack of validated and sufficiently controlled clinical trials presents a challenge for phage therapy for clinical use. Planning and designing pharmacological aspects and dosages are major activities to consider for clinical use.84 The dissemination of phages in the body may reduce their efficacy because phages require direct contact with bacteria at an optimum concentration to effectively act on the bacteria. Topical applications and many other methods are used to administer phages. Phage therapy can be given as monotherapy, combination therapy, or phage cocktails; however, the last of these provides broad-spectrum activity and a low risk of the development of resistance. More importantly, combination therapy largely elevates the challenge of diagnosing inflammatory effects, the potential for gene transfer, and phage resistance development for all phages in the cocktail.85 Another vital consideration to be addressed before any clinical trial is the onset of toxic shock, as phages are bactericidal.86
Clinical Trials Involving Phages
Clinical trials on phage therapy practices in Georgia and Poland were discussed by Kutter et al87 and are prominently mentioned in many studies in the literature, confirming the safety of phages in treating venous leg ulcers88 and their safety and efficacy in chronic otitis.89 Rhoads et al reported no adverse side effects in a patient with venous leg ulcers in a small phase I clinical trial on phage therapy.88 The efficacy and safety of anti-pseudomonal phages in late-stage recurrent otitis, which was mainly controlled by MDR P. aeroginosa, were demonstrated by Wright et al.89 Although phage therapy is incorporated in the health policies of Eastern European countries, the above-mentioned controlled clinical trials were among the first trials conducted in humans in the Western world. Currently, many clinical trials have been registered.88,89
Scientifically sound clinical trials are essential for phage therapy to be accepted by the Western clinical world. Although many observational phage therapy studies have been conducted and were effective, they have limitations owing to small sample sizes and poor control. In addition, despite the existence of promising case studies, strong clinical trial data are expected by regulators to prepare guidelines for phage therapy81 to be approved by the United States Food and Drug Administration (FDA).
Determinants of Human Phage Therapy Trials
Virulent Genes
The pathogenicity and severity of a disease may increase when virulent genes are acquired from other virulent species, which may cause treatment failure.90 Phages can carry virulent genes that may increase the virulence and pathogenicity of bacteria during lysogenization. Phage virulent genes are detected in many human pathogens, including E. coli, P. aeroginosa, S. aureus, and S. pyogenes. Currently, not all phage-encoded virulent genes have been identified. Therefore, it is vital that the metagenome and each genome sequence are added to databases of bacterial virulent genes and antibiotic resistance genes to ensure safety.91,92
Transduction
Bacteria can acquire virulent and antibiotic-resistant genes through phage-mediated transduction. This can be mitigated by avoiding known transducing phages.93
Disturbance of Commensal Microbiota
It is crucial to know about and investigate all interactions of phages with human niche microbiota when using phages as therapeutic agents, and specifically any possible perturbations in the human microbiota due to strong selective pressure of lytic phages. Additional mechanistic investigations that clearly show the nature of host–phage dynamics in niche microbiota will help to hasten the process of obtaining regulatory approval for phage therapy in Western medicine.94 This is sometimes mentioned as a concern. However, phages are generally species specific, so that disturbance of the commensal microbiota is minimal.95
Quality and Safety Requirements
The safety and quality of phage preparations will determine the success of phage therapy. The production of phages for therapy must follow strict regulations to assure their quality for their intended use, even if no clear guidelines have been established for the manufacture of phages.96 The presence of impurities such as endotoxins must be avoided or kept below the threshold in phage preparations, but none has reached the optimal level so far. The quality of phage therapy is regulated by assessing and checking their stability, sterility, and cytotoxicity, and performing pH measurements regularly.97
Phage Storage and Handling
Phages for therapy are mainly in the form of water suspension and are freshly made. There is not enough understanding of how to process phages with well-defined pharmaceutical properties and stability. Phages lack stability, which is one of the ideal characteristics of drugs, along with specificity, high affinity, solubility, and safety.98,99 They are partially stable in solution owing to their protein structure. The structural instability of phages makes long-term storage difficult and cooling is essential to store phage preparations. Phages in aqueous solution can be stabilized by the addition of stability enhancers or by processing them into another formulation, through lyophilization, spray drying, or incorporating them into ointments, biodegradable polymer matrices, or microparticles.100–102
Conclusion and Perspectives
Current challenges facing phage therapy are: compatibility with current quality and safety requirements, necessitating the stability of phage preparations for long periods of time, designing effective assays for phage screening, overcoming the limited activity of phages in biofilms, controlling and disabling the appearance of bacterial resistance to phages, production of antibody-neutralizing phages, sequestration by the spleen and liver, and launching an appropriate regulatory framework for phage products.31,103 A comparison of different antibacterial treatment mechanisms by administering antibiotics and phages is shown in Table 1. The pros and cons of phage therapy are shown in Table 2.
 |
Table 1 Comparison of Antibiotic and Lytic Phage Therapy |
 |
Table 2 Pros and Cons of Lytic and Temperate Phage Therapy |
Phage therapy is a promising approach to combat bacterial infections, including multidrug-resistant bacteria. For efficacious phage therapy, phages need to be present in high concentrations, stable, able to encounter bacteria with no restrictions, and able to replicate. Phage therapy can be used either as an alternative or as a supplement to antibiotics. The increment of antibiotic-resistant bacteria can be reduced using phage cocktails, phage-derived lytic proteins, bioengineered phages, and/or antibiotics. Phage therapy is highly specific, effective in lysing the targeted bacteria, safe (as seen in Eastern Europe), and quickly modifiable to fight newly emerging bacterial threats. Although it has been shown to be effective in some clinical trials, many of the trials do not meet the existing high standards for clinical trials and many questions remain regarding the therapeutic use of phages. Better understanding of phage–host and phage–human interactions, phage diversity, phage dynamics, and genome function is essential to develop a new strategy in the fight against bacterial infections and to overcome the challenges associated with phage therapy. Nevertheless, despite numerous studies having been conducted on phage therapy in the last few decades in the Western world, and before that in Eastern Europe, no phage therapies for humans have been approved by the European Union or the US FDA. Finally, further all-inclusive intensive studies are necessary to justify phage therapy for large-scale clinical use.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that there are no conflicts of interest in this work.
References
1. Danis-Wlodarczyk K, Dąbrowska K, Abedon ST. Phage therapy: the pharmacology of antibacterial viruses. Curr Issues Mol Biol. 2020;40:81–164. doi:10.21775/cimb.040.081
2. Hendrix RW. Bacteriophages: evolution of the majority. Theor Popul Biol. 2002;61(4):471–480. doi:10.1006/tpbi.2002.1590
3. Sabour PM, Griffiths MW. Bacteriophages in the Control of Food-and Waterborne Pathogens. American Society for Microbiology Press; 2010.
4. Servick K. Beleaguered phage therapy trial presses on. American Association for the Advancement of Science; 2016.
5. Viertel TM, Ritter K, Horz H-P. Viruses versus bacteria—novel approaches to phage therapy as a tool against multidrug-resistant pathogens. J Antimicrob Chemother. 2014;69(9):2326–2336. doi:10.1093/jac/dku173
6. Lin DM, Koskella B, Lin HC. Phage therapy: an alternative to antibiotics in the age of multi-drug resistance. World J Gastrointest Pharmacol Ther. 2017;8(3):162. doi:10.4292/wjgpt.v8.i3.162
7. Patey O, McCallin S, Mazure H, Liddle M, Smithyman A, Dublanchet A. Clinical indications and compassionate use of phage therapy: personal experience and literature review with a focus on osteoarticular infections. Viruses. 2019;11(1):18. doi:10.3390/v11010018
8. Love MJ, Bhandari D, Dobson RC, Billington C. Potential for bacteriophage endolysins to supplement or replace antibiotics in food production and clinical care. Antibiotics. 2018;7(1):17. doi:10.3390/antibiotics7010017
9. Drulis-Kawa Z, Majkowska-Skrobek G, Maciejewska B. Bacteriophages and phage-derived proteins–application approaches. Curr Med Chem. 2015;22(14):1757–1773. doi:10.2174/0929867322666150209152851
10. Hobbs Z, Abedon ST, Millard A. Diversity of phage infection types and associated terminology: the problem with ‘Lytic or lysogenic’. FEMS Microbiol Lett. 2016;363(7):fnw047. doi:10.1093/femsle/fnw047
11. Chen Y, Yang L, Yang D, et al. Specific integration of temperate phage decreases the pathogenicity of host bacteria. Front Cell Infect Microbiol. 2020;10:14. doi:10.3389/fcimb.2020.00014
12. Örmälä A-M, Jalasvuori M. Phage therapy: should bacterial resistance to phages be a concern even in the long run?. Bacteriophage. 2013;3(1):e24219. doi:10.4161/bact.24219
13. Brives C, Pourraz J. Phage therapy as a potential solution in the fight against AMR: obstacles and possible futures. Palgrave Commun. 2020;6(1):1–11. doi:10.1057/s41599-020-0478-4
14. Reuter M, Kruger DH. Approaches to optimize therapeutic bacteriophage and bacteriophage-derived products to combat bacterial infections. Virus Genes. 2020;56(2)1–14.
15. Romero-Calle D, Benevides RG, Góes-Neto A, Billington C. Bacteriophages as alternatives to antibiotics in clinical care. Antibiotics. 2019;8(3):138. doi:10.3390/antibiotics8030138
16. Pirnay J-P. Phage therapy in the year 2035. Front Microbiol. 2020;11:1171. doi:10.3389/fmicb.2020.01171
17. Jończyk-Matysiak E, Weber-Dąbrowska B, Owczarek B. Phage-phagocyte interactions and their implications for phage application as therapeutics. Viruses. 2017;9(6):150. doi:10.3390/v9060150
18. Górski A, Dąbrowska K, Międzybrodzki R. Phages and immunomodulation. Future Microbiol. 2017;12(10):905–914. doi:10.2217/fmb-2017-0049
19. Krut O, Bekeredjian-Ding I. Contribution of the immune response to phage therapy. J Immunol. 2018;200(9):3037–3044. doi:10.4049/jimmunol.1701745
20. Cafora M, Brix A, Forti F, et al. Phages as immunomodulators and their promising use as anti-inflammatory agents in a CFTR loss-of-function zebrafish model. J Cyst Fibros. 2021;20(6):1046–1052. doi:10.1016/j.jcf.2020.11.017
21. Hashiguchi S, Yamaguchi Y, Takeuchi O, Akira S, Sugimura K. Immunological basis of M13 phage vaccine: regulation under MyD88 and TLR9 signaling. Biochem Biophys Res Commun. 2010;402(1):19–22. doi:10.1016/j.bbrc.2010.09.094
22. Dąbrowska K, Miernikiewicz P, Piotrowicz A. Immunogenicity studies of proteins forming the T4 phage head surface. J Virol. 2014;88(21):12551–12557. doi:10.1128/JVI.02043-14
23. Van Belleghem JD, Vaneechoutte M, Barr JJ, Bollyky PL, Bollyky P. Interactions between bacteriophage, bacteria, and the mammalian immune system. Viruses. 2018;11(1):10. doi:10.3390/v11010010
24. Gã³rski A, Miä™dzybrodzki R, Borysowski J, et al. Phage as a modulator of immune responses: practical implications for phage therapy. Adv Virus Res. 2012;83:41–71.
25. Szermer-Olearnik B, Boratyå„ski J, Skurnik M. Removal of endotoxins from bacteriophage preparations by extraction with organic solvents. PLoS One. 2015;10(3):e0122672. doi:10.1371/journal.pone.0122672
26. Leung CYJ, Weitz JS. Modeling the synergistic elimination of bacteria by phage and the innate immune system. J Theor Biol. 2017;429:241–252. doi:10.1016/j.jtbi.2017.06.037
27. Wang J, Hu B, Xu M, et al. Therapeutic effectiveness of bacteriophages in the rescue of mice with extended spectrum β-lactamase-producing Escherichia coli bacteremia. Int J Mol Med. 2006;17(2):347–355.
28. Biswas B, Adhya S, Washart P, et al. Bacteriophage therapy rescues mice bacteremic from a clinical isolate of vancomycin-resistant Enterococcus faecium. Infect Immun. 2002;70(1):204–210. doi:10.1128/IAI.70.1.204-210.2002
29. Hodyra-Stefaniak K, Miernikiewicz P, Drapała J. Mammalian Host-Versus-Phage immune response determines phage fate in vivo. Sci Rep. 2015;5(1):1–13. doi:10.1038/srep14802
30. Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8(5):317–327. doi:10.1038/nrmicro2315
31. Pires DP, Costa AR, Pinto G, Meneses L, Azeredo J. Current challenges and future opportunities of phage therapy. FEMS Microbiol Rev. 2020;44(6):684–700. doi:10.1093/femsre/fuaa017
32. Weissman JL, Holmes R, Barrangou R, et al. Immune loss as a driver of coexistence during host-phage coevolution. ISME J. 2018;12(2):585–597. doi:10.1038/ismej.2017.194
33. Koskella B, Brockhurst MA. Bacteria–phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol Rev. 2014;38(5):916–931. doi:10.1111/1574-6976.12072
34. Azam AH, Tanji Y. Bacteriophage-host arm race: an update on the mechanism of phage resistance in bacteria and revenge of the phage with the perspective for phage therapy. Appl Microbiol Biotechnol. 2019;103(5):2121–2131. doi:10.1007/s00253-019-09629-x
35. Burmeister AR, Turner PE. Trading-off and trading-up in the world of bacteria–phage evolution. Current Biol. 2020;30(19):R1120–R1124. doi:10.1016/j.cub.2020.07.036
36. Vasu K, Nagaraja V. Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol Mol Biol Rev. 2013;77(1):53–72. doi:10.1128/MMBR.00044-12
37. Dryden D. S-Adenosylmethionine-Dependent Methyltransferases: Structures and Functions. Bacterial DNA Methyltransferases. Singapore: World Scientific Publishing; 1999:283–340.
38. Bonsma-Fisher M, Soutière D, Goyal S. How adaptive immunity constrains the composition and fate of large bacterial populations. Proc Natl Acad Sci. 2018;115(32):E7462–E7468. doi:10.1073/pnas.1802887115
39. Bennett P. Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. Br J Pharmacol. 2008;153(S1):S347–S57. doi:10.1038/sj.bjp.0707607
40. Dahlberg C, Chao L. Amelioration of the cost of conjugative plasmid carriage in Escherichia coli K12. Genetics. 2003;165(4):1641–1649. doi:10.1093/genetics/165.4.1641
41. Dionisio F, Conceicao I, Marques A, Fernandes L, Gordo I. The evolution of a conjugative plasmid and its ability to increase bacterial fitness. Biol Lett. 2005;1(2):250–252. doi:10.1098/rsbl.2004.0275
42. Malone LM, Birkholz N, Fineran PC. Conquering CRISPR: how phages overcome bacterial adaptive immunity. Curr Opin Biotechnol. 2020;68:30–36. doi:10.1016/j.copbio.2020.09.008
43. Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. doi:10.1126/science.1138140
44. Deveau H, Barrangou R, Garneau JE, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190(4):1390–1400. doi:10.1128/JB.01412-07
45. Semenova E, Jore MM, Datsenko KA, et al. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci. 2011;108(25):10098–10103. doi:10.1073/pnas.1104144108
46. Shearer WT, Lugg DJ, Rosenblatt HM, et al. Antibody responses to bacteriophage φX-174 in human subjects exposed to the Antarctic winter-over model of spaceflight. J Allergy Clin Immunol. 2001;107(1):160–164. doi:10.1067/mai.2001.112269
47. Rubinstein A, Mizrachi Y, Bernstein L, et al. Progressive specific immune attrition after primary, secondary and tertiary immunizations with bacteriophage φX174 in asymptomatic HIV-1 infected patients. AIDS. 2000;14(4):F55–F62. doi:10.1097/00002030-200003100-00004
48. Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun. 2012;3(1):1–7. doi:10.1038/ncomms1937
49. Swarts DC, Mosterd C, Van Passel MW, Brouns SJ, Mokrousov I. CRISPR interference directs strand specific spacer acquisition. PLoS One. 2012;7(4):e35888. doi:10.1371/journal.pone.0035888
50. Pyenson NC, Gayvert K, Varble A, Elemento O, Marraffini LA. Broad targeting specificity during bacterial type III CRISPR-Cas immunity constrains viral escape. Cell Host Microbe. 2017;22(3):343–53. e3. doi:10.1016/j.chom.2017.07.016
51. Marino ND, Pinilla-Redondo R, Csörgő B, Bondy-Denomy J. Anti-CRISPR protein applications: natural brakes for CRISPR-Cas technologies. Nat Methods. 2020;17(5):471–479. doi:10.1038/s41592-020-0771-6
52. Wiegand T, Karambelkar S, Bondy-Denomy J, Wiedenheft B. Structures and strategies of anti-CRISPR-mediated immune suppression. Annu Rev Microbiol. 2020;74:21–37. doi:10.1146/annurev-micro-020518-120107
53. Patterson AG, Yevstigneyeva MS, Fineran PC. Regulation of CRISPR–Cas adaptive immune systems. Curr Opin Microbiol. 2017;37:1–7. doi:10.1016/j.mib.2017.02.004
54. Borges AL, Castro B, Govindarajan S, Solvik T, Escalante V, Bondy-Denomy J. Bacterial alginate regulators and phage homologs repress CRISPR–Cas immunity. Nature Microbiol. 2020;5(5):679–687. doi:10.1038/s41564-020-0691-3
55. Hargreaves KR, Kropinski AM, Clokie MR. Bacteriophage behavioral ecology: how phages alter their bacterial host’s habits. Bacteriophage. 2014;4(3):e85131. doi:10.4161/bact.29866
56. Patterson AG, Jackson SA, Taylor C, et al. Quorum sensing controls adaptive immunity through the regulation of multiple CRISPR-Cas systems. Mol Cell. 2016;64(6):1102–1108. doi:10.1016/j.molcel.2016.11.012
57. Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2013;493(7432):429–432. doi:10.1038/nature11723
58. Oechslin F. Resistance development to bacteriophages occurring during bacteriophage therapy. Viruses. 2018;10(7):351. doi:10.3390/v10070351
59. Chan BK, Abedon ST, Loc-Carrillo C. Phage cocktails and the future of phage therapy. Future Microbiol. 2013;8(6):769–783. doi:10.2217/fmb.13.47
60. Pirnay J-P, De Vos D, Verbeken G, et al. The phage therapy paradigm: pret-a-porter or sur-mesure? Pharm Res. 2011;28(4):934–937. doi:10.1007/s11095-010-0313-5
61. Górski A, Międzybrodzki R, Weber-Dąbrowska B, et al. Phage therapy: combating infections with potential for evolving from merely a treatment for complications to targeting diseases. Front Microbiol. 2016;7:1515. doi:10.3389/fmicb.2016.01515
62. Lu TK, Collins JJ. Dispersing biofilms with engineered enzymatic bacteriophage. Proc Natl Acad Sci. 2007;104(27):11197–11202. doi:10.1073/pnas.0704624104
63. Tinoco JM, Buttaro B, Zhang H, Liss N, Sassone L, Stevens R. Effect of a genetically engineered bacteriophage on Enterococcus faecalis biofilms. Arch Oral Biol. 2016;71:80–86. doi:10.1016/j.archoralbio.2016.07.001
64. Tinoco JM, Buttaro B, Zhang H, Liss N, Sassone L, Stevens R. Effect of a genetically engineered bacteriophage on Enterococcus faecalis biofilms. Arch Oral Biol. 2019;71:80–86.
65. Monteiro R, Pires DP, Costa AR, Azeredo J. Phage therapy: going temperate? Trends Microbiol. 2019;27(4):368–378. doi:10.1016/j.tim.2018.10.008
66. Ando H, Lemire S, Pires DP, Lu TK. Engineering modular viral scaffolds for targeted bacterial population editing. Cell Systems. 2015;1(3):187–196. doi:10.1016/j.cels.2015.08.013
67. Lu TK, Collins JJ. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc Natl Acad Sci. 2009;106(12):4629–4634. doi:10.1073/pnas.0800442106
68. Yacoby I, Bar H, Benhar I. Targeted drug-carrying bacteriophages as antibacterial nanomedicines. Antimicrob Agents Chemother. 2007;51(6):2156–2163. doi:10.1128/AAC.00163-07
69. Russel M, Linderoth NA, Šali A. Filamentous phage assembly: variation on a protein export theme. Gene. 1997;192(1):23–32. doi:10.1016/S0378-1119(96)00801-3
70. Kortright KE, Chan BK, Koff JL, Turner PE. Phage therapy: a renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe. 2019;25(2):219–232. doi:10.1016/j.chom.2019.01.014
71. Wang N, Deaton J, Young R. Sizing the holin lesion with an endolysin-β-galactosidase fusion. J Bacteriol. 2003;185(3):779–787. doi:10.1128/JB.185.3.779-787.2003
72. Jervis EJ, Guarna MM, Doheny JG, Haynes CA, Kilburn DG. Dynamic localization and persistent stimulation of factor‐dependent cells by a stem cell factor/cellulose binding domain fusion protein. Biotechnol Bioeng. 2005;91(3):314–324. doi:10.1002/bit.20611
73. Adhya S, Merril CR, Biswas B. Therapeutic and prophylactic applications of bacteriophage components in modern medicine. Cold Spring Harb Perspect Med. 2014;4(1):a012518. doi:10.1101/cshperspect.a012518
74. Lukacik P, Barnard TJ, Keller PW, et al. Structural engineering of a phage lysin that targets Gram-negative pathogens. Proc Natl Acad Sci. 2012;109(25):9857–9862. doi:10.1073/pnas.1203472109
75. Yosef I, Kiro R, Molshanski-Mor S, Edgar R, Qimron U. Different approaches for using bacteriophages against antibiotic-resistant bacteria. Bacteriophage. 2014;4(1):19549–19554. doi:10.4161/bact.28491
76. Roach DR, Donovan DM. Antimicrobial bacteriophage-derived proteins and therapeutic applications. Bacteriophage. 2015;5(3):e1062590. doi:10.1080/21597081.2015.1062590
77. Yosef I, Manor M, Kiro R, Qimron U. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc Natl Acad Sci. 2015;112(23):7267–7272. doi:10.1073/pnas.1500107112
78. Hodyra-Stefaniak K, Miernikiewicz P, Drapała J, et al. Mammalian Host-Versus-Phage immune response determines phage fate in vivo. Sci Rep. 2015;5(1):1–13.
79. Chopra S, Harjai K, Chhibber S. Potential of combination therapy of endolysin MR-10 and minocycline in treating MRSA induced systemic and localized burn wound infections in mice. Int J Med Microbiol. 2016;306(8):707–716. doi:10.1016/j.ijmm.2016.08.003
80. Wittekind M, Schuch R. Cell wall hydrolases and antibiotics: exploiting synergy to create efficacious new antimicrobial treatments. Curr Opin Microbiol. 2016;33:18–24. doi:10.1016/j.mib.2016.05.006
81. Furfaro LL, Payne MS, Chang BJ. Bacteriophage therapy: clinical trials and regulatory hurdles. Front Cell Infect Microbiol. 2018;8:376. doi:10.3389/fcimb.2018.00376
82. Harper DR. Criteria for selecting suitable infectious diseases for phage therapy. Viruses. 2018;10(4):177. doi:10.3390/v10040177
83. de Jonge PA, Nobrega FL, Brouns SJ, Dutilh BE. Molecular and evolutionary determinants of bacteriophage host range. Trends Microbiol. 2018;27(1):51–63. doi:10.1016/j.tim.2018.08.006
84. Payne RJ, Jansen VA. Pharmacokinetic principles of bacteriophage therapy. Clin Pharmacokinet. 2003;42(4):315–325. doi:10.2165/00003088-200342040-00002
85. Parracho HM, Burrowes BH, Enright MC, McConville ML, Harper DR. The role of regulated clinical trials in the development of bacteriophage therapeutics. J Mol Genetic Med. 2012;6:279. doi:10.4172/1747-0862.1000050
86. Speck P, Smithyman A, Millard A. Safety and efficacy of phage therapy via the intravenous route. FEMS Microbiol Lett. 2016;363(3):fnv242. doi:10.1093/femsle/fnv242
87. Kutter E, De Vos D, Gvasalia G, et al. Phage therapy in clinical practice: treatment of human infections. Curr Pharm Biotechnol. 2010;11(1):69–86. doi:10.2174/138920110790725401
88. Rhoads D, Wolcott R, Kuskowski M, Wolcott B, Ward L, Sulakvelidze A. Bacteriophage therapy of venous leg ulcers in humans: results of a phase I safety trial. J Wound Care. 2009;18(6):237–243. doi:10.12968/jowc.2009.18.6.42801
89. Wright A, Hawkins C, Änggã¥rd E, Harper D. A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin otolaryngol. 2009;34(4):349–357. doi:10.1111/j.1749-4486.2009.01973.x
90. Abd El-Baky RM, Ibrahim RA, Mohamed DS, Ahmed EF, Hashem ZS. Prevalence of virulence genes and their association with antimicrobial resistance among pathogenic E. coli isolated from Egyptian patients with different clinical infections. Infect Drug Resist. 2020;13:1221. doi:10.2147/IDR.S241073
91. Yang J, Chen L, Sun L, Yu J, Jin Q. VFDB 2008 release: an enhanced web-based resource for comparative pathogenomics. Nucleic Acids Res. 2007;36(suppl_1):D539–D542. doi:10.1093/nar/gkm951
92. Sarker SA, McCallin S, Barretto C, et al. Oral T4-like phage cocktail application to healthy adult volunteers from Bangladesh. Virology. 2012;434(2):222–232. doi:10.1016/j.virol.2012.09.002
93. Muniesa M, Imamovic L, Jofre J. Bacteriophages and genetic mobilization in sewage and faecally polluted environments. Microb Biotechnol. 2011;4(6):725–734. doi:10.1111/j.1751-7915.2011.00264.x
94. Divya Ganeshan S, Hosseinidoust Z. Phage therapy with a focus on the human microbiota. Antibiotics. 2019;8(3):131. doi:10.3390/antibiotics8030131
95. Santos S, Fernandes E, Carvalho CM, et al. Selection and characterization of a multivalent Salmonella phage and its production in a nonpathogenic Escherichia coli strain. Appl Environ Microbiol. 2010;76(21):7338–7342. doi:10.1128/AEM.00922-10
96. Mutti M, Corsini L, Zhou H. Robust approaches for the production of active ingredient and drug product for human phage therapy. Front Microbiol. 2019;10:10. doi:10.3389/fmicb.2019.00010
97. Pirnay J-P, Blasdel BG, Bretaudeau L, et al. Quality and safety requirements for sustainable phage therapy products. Pharm Res. 2015;32(7):2173–2179. doi:10.1007/s11095-014-1617-7
98. Vandenheuvel D, Lavigne R, Brüssow H. Bacteriophage therapy: advances in formulation strategies and human clinical trials. Ann rev virol. 2015;2:599–618. doi:10.1146/annurev-virology-100114-054915
99. Gill JJ, Hyman P. Phage choice, isolation, and preparation for phage therapy. Curr Pharm Biotechnol. 2010;11(1):2–14. doi:10.2174/138920110790725311
100. Ackermann H-W, Tremblay D, Moineau S. Long-term bacteriophage preservation; 2004.
101. Tovkach F, Zhuminska G, Kushkina A. Long-term preservation of unstable bacteriophages of enterobacteria. Mikrobiol Z. 2012;74(2):60–66.
102. Jończyk E, Kłak M, Międzybrodzki R, Górski A. The influence of external factors on bacteriophages. Folia Microbiol. 2011;56(3):191–200. doi:10.1007/s12223-011-0039-8
103. Pires DP, Boas DV, Sillankorva S, Azeredo J, Goff SP. Phage therapy: a step forward in the treatment of Pseudomonas aeruginosa infections. J Virol. 2015;89(15):7449–7456. doi:10.1128/JVI.00385-15
104. Adesanya O, Oduselu T, Akin-Ajani O, Adewumi OM, Ademowo OG. An exegesis of bacteriophage therapy: an emerging player in the fight against anti-microbial resistance. AIMS Microbiol. 2014;6(3):204. doi:10.3934/microbiol.2020014
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
