Back to Journals » Journal of Blood Medicine » Volume 14
Prevalence of Gene Rearrangement by Multiplex PCR in De Novo Acute Myeloid Leukemia in Adult Iraqi Patients
Authors AlJabban A, Alalsaidissa J
Received 7 May 2023
Accepted for publication 1 August 2023
Published 11 August 2023 Volume 2023:14 Pages 445—453
DOI https://doi.org/10.2147/JBM.S416825
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Ali AlJabban, Jaffar Alalsaidissa
Department of Pathology, College of Medicine, University of Baghdad, Baghdad, Iraq
Correspondence: Ali AlJabban, Department of Pathology, College of medicine, University of Baghdad, Baghdad, Iraq, Tel +9647706056226, Email [email protected]
Introduction: Gene rearrangements of acute myeloid leukemia (AML) play a significant role in categorizing patients and provide valuable information about prognosis and treatment choices. However, in Iraq, the prevalence and prognostic significance of gene rearrangements in AML have not been previously examined.
Methods: This study utilized a multiplex reverse transcription real-time PCR (RT-qPCR) system to identify gene rearrangements in a group of 115 adult patients from Iraq who had been diagnosed with De Novo AML. The diagnosis of AML was confirmed through blood film and flow cytometry. The ethical committee of the College of Medicine at the University of Baghdad provided approval for this research study.
Results: In this study, 66.1% of the patients diagnosed with acute myeloid leukemia (AML) exhibited distinct genetic abnormalities. Among these abnormalities, the most frequent was the rearrangement involving the KMT2A gene, observed in 19.9% of the patients. The risk stratification analysis revealed that 40% of the patients were classified as having a favorable risk, 4.3% as intermediate risk, and 25.2% as adverse risk. A subtype of AML known as core-binding factor (CBF) AML was identified in 21.7% of the cases, with 84% of these patients achieving complete remission. The NPM-RARA gene rearrangement, found in 43% of acute promyelocytic leukemia (APL) cases, was associated with a 71% complete remission rate. Among patients with KMT2A rearrangement, which accounted for 19.9% of all AML cases, the MLL-AF10 rearrangement was the most common, although only one patient with KMT2A rearrangement achieved complete remission. Furthermore, the analysis of demographic data revealed a significant association between increased risk and advanced age, presence of comorbidities, and FAB classification (M0 subtype).
Conclusion: The prevalence of genetic rearrangements in Iraqi De Novo AML patients is higher than the global trend, highlighting the importance of genetic characterization in risk assessment and treatment decisions.
Keywords: gene rearrangement, acute myeloid leukemia, APL, core-binding factor, KMT2A rearrangement
Introduction
Over the past two decades, the classification of acute myeloid leukemia (AML) has undergone significant revisions, shifting towards a greater reliance on the understanding of its genetic landscape. This revolution encompasses various aspects, including chromosomal abnormalities, recurrent gene mutations, and deregulation of microRNAs,1 which have provided valuable insights into prognosis and treatment response prediction.2 The European Leukemia Network (ELN) and National Comprehensive Cancer Network (NCCN) guidelines now recommend comprehensive genetic testing for all newly diagnosed AML patients.3,4 This testing includes the screening of gene rearrangements, as it is considered a fundamental requirement for accurately assessing the genetic profile of patients. However, in Iraq, the prevalence and prognostic value of gene rearrangements have not been previously investigated comprehensively. Therefore, the aim of our study is to evaluate the occurrence of gene rearrangements in De Novo AML patients in Iraq and examine their correlation with risk stratification and clinicopathological factors.
Materials and Methods
A prospective cohort study was conducted at Baghdad teaching hospital, involving the recruitment of 115 adult patients who were diagnosed with De Novo AML. The diagnosis of AML was confirmed using blood film and flow cytometry. Data collection was carried out between December 2020 and May 2022. The study was approved by the ethical committee of the College of Medicine, University of Baghdad, and informed consent was obtained from all participating patients.
The inclusion criteria for the study were as follows:
- Patients diagnosed with AML (confirmed by blood film and flow cytometry) within one week after treatment initiation, including newly diagnosed cases.
- Patients aged 18 years and above.
The following were the exclusion criteria:
- Patients who underwent Allogeneic Transplantation.
- Pregnant women.
- Patients with a history of cancer (CA) or myelodysplastic syndrome (MDS).
Method
Peripheral blood samples (3 mL) were obtained from eligible patients using EDTA tubes to ensure sample freshness. RNA extraction was performed using the PAXgene Blood RNA System, which consists of a blood collection tube (PAXgene Blood RNA Tube, BRT) and a nucleic acid purification kit (PAXgene Blood RNA Kit). A comprehensive RT-qPCR-based assay (Leukemia Q-Fusion Screening Kit, Zeesan Biotech Co., Ltd) was utilized to detect more than 140 breakpoints for 30 fusion genes, including MLL-AF9, PML-RARA, AML-ETO, MLL-AF4, TEL-AML1, E2A-PBX1, MLL-ENL, BCR-ABL1, SIL-TAL1, MLL-AF10, CBF-MYH11, AML1-MDS1/EV11, FIP1L1-PDGFRA, SET-CAN, E2A-HLF, DEK-CAN, MLL-SEPT6, TLS-ERG, TEL-PDGFRB, MLL-ELL, MLL-AF17, NPM-RARA, NPM-MLF1, PLZF-RARA, MLL-AF1q, MLL-AF1P, TEL-ABL, AML1-MTG16, AML1-EAP, and MLL-AF6.
After RNA extraction, the extracted RNA was combined with pre-made cDNA from the kit. The resulting mixture was divided into eight tubes, each containing specific PCR primers and probes designed for the detection of fusion genes. Real-time qPCR was performed using optical filters to capture fluorescence signals (FAM, HEX, ROX, and Cy5). Samples with a threshold cycle (Ct) value above 25 for Cy5 were considered of low quality and were retested. Samples with a Ct value above 30 were considered to exhibit non-specific amplification and were also repeated. Samples displaying signals with a Ct value below 30 (excluding Cy5) were considered positive for the presence of fusion genes. Furthermore, information regarding NPM1 and FLT-3 mutations was obtained from the patients’ data.
Ethical Consideration
The study received approval from the scientific committee department of Pathology at the College of Medicine, University of Baghdad, to proceed with the research in compliance with Declaration of Helsinki. Informed consent was obtained from all participants prior to their involvement in the study.
Results
The study included patients with a mean age of 45.1 ± 17.5 years, ranging from 18 to 84 years. The majority of the participants were female (59.1%), and only 16.5% reported being smokers. Among the patients, 2.6% had hypertension, while 7.8% had both hypertension and diabetes mellitus (DM). Splenomegaly was observed in 27.8% of the patients, while hepatomegaly was present in 17.4% of the cases (Table 1).
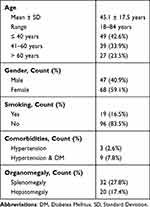 |
Table 1 Demographic Characteristics of Included Patients (n=115) |
Gene Rearrangement
The gene rearrangement showed that acute myeloid leukemia with defining genetic abnormalities presented in 66.1% of patients. Among these abnormalities, the most frequent was AML with KMT2A rearrangement, accounting for 19.9% of cases. Differentiation-based classification of AML demonstrated that acute myelomonocytic leukemia was the most prevalent subtype, observed in 10 patients (8.7%) (Table 2). The NPM1 was presented in 33 (28.7%) patients, and FLT-3 was presented in 18 (15.7%) patients.
 |
Table 2 Prevalence of AML Defining Cytogenetics Abnormalities in Study Cohort According to WHO 2022 Classification |
Applying the European Leukemia Network (ELN) risk stratification for cytogenetics, the distribution of risk categories was as follows: 25 patients with favorable risk, 5 patients with intermediate risk, and 25 patients with adverse risk. Additionally, considering the presence of the PML-RARA fusion, which is associated with a good prognosis, 9 patients were included in the favorable prognosis group. Patients classified as acute myeloid leukemia with differentiation were excluded from this analysis, as their abnormal karyotyping was not assessed in this study. Patients with FLT-3 mutation alone were categorized as adverse risk, while those with FLT-3 mutation and other cytogenetic abnormalities followed the cytogenetic risk stratification.5 Overall, the study revealed that 46 patients (40%) were classified as favorable risk, 5 patients (4.3%) as intermediate risk, and 29 patients (25.2%) as adverse risk (Table 3).
 |
Table 3 Relationship of Risk Stratification and Response to Treatment (All Studied Group) |
Follow-Up Period and Response
Each patient was monitored for a duration of one month starting from day 0 of receiving standardized chemotherapy. For statistical purposes, cases of complete remission with incomplete hematological recovery (CRi), five cases, were combined with complete remission (CR). Among the study cohort, a total of 71 (61.74%) patients achieved complete remission, indicating a positive response to treatment. On the other hand, 32 patients (27.8%) experienced treatment failure, characterized by persistence of disease (BP). Unfortunately, there were 12 patients (10.4%) who died before completing the first month of treatment. Out of these, 10 patients refused the treatment, while the remaining two patients passed away within one week of diagnosis.
A significant association was observed between treatment response and risk stratification, indicating that a deterioration in treatment response correlated with a higher risk group (p=0.0001) (Table 3). The analysis of demographic data revealed several significant associations. Firstly, advanced age was significantly associated with a higher proportion of adverse risk cases. Secondly, the presence of comorbidities was also significantly associated with adverse risk. Additionally, specific AML subtypes, namely AML-M0, AML-M4, and AML-M5, were significantly associated with adverse risk. Lastly, there was a significant association between the presence of splenomegaly and risk stratification (Table 4).
 |
Table 4 Relationship of Demographic Data with Risk Stratification for All Patients |
Core-Binding Factor (CBF) Acute Myeloid Leukemia (AML)
Core-binding factor (CBF) AML is a specific subtype of AML characterized by two distinct genetic abnormalities: t(8;21)(q22;q22) or inv(16)(p13q22)/t(16;16). These abnormalities lead to the formation of two fusion genes: RUNX1/RUNX1T1 (also known as AML1/ETO) and CBFB/MYH11. CBF-AML is known to be associated with a favorable treatment response.
In our analysis, CBF-AML was observed in 21.7% of the patients. Among patients with CBF-AML, 84% achieved complete remission, indicating a positive response to treatment. Only 4 patients (16%) experienced treatment failure, as evidenced by persistent disease (BP). Importantly, no deaths were reported among the patients with CBF AML during the study period.
Acute Promyelocytic Leukemia
Among the patients included in the study, 16 individuals (13.9%) were diagnosed with Acute Promyelocytic Leukemia (APL). The diagnosis of APL was based on the presence of RARA translocations, with PML-RARA representing 37% of APL cases and NPM-RARA representing 43% of APL cases (all patients with NPM-RARA also had NPM1 mutation). Additionally, PLZF-RARA was observed in 3 cases (out of 16 APL cases).
There were significant differences observed in the treatment response among patients with APL based on their specific gene rearrangements. For patients with PML-RARA, all of them achieved complete remission (CR) as a response to treatment and among patients with NPM-RARA, a 71% complete remission rate was observed. In contrast, among the three patients with PLZF-RARA, two of them unfortunately died and one experienced treatment failure (BP).
Furthermore, there was a significant difference in treatment response, indicating that patients with PLZF-RARA had a worsened response compared to individuals with other gene rearrangements.
Acute Myeloid Leukemia with KMT2A Rearrangement
Acute myeloid leukemia with KMT2A rearrangement represented in 19.9% of patients. Among the various KMT2A rearrangements observed, MLL-AF10 was the most common (Table 2). However, the response to treatment in patients with KMT2A rearrangement was found to be inadequate. Among these patients, 65% experienced treatment failure (BP), 30% died, and only one patient achieved complete remission (CR).
Notably, there was no significant difference observed in the treatment response between patients with different KMT2A rearrangements. However, when comparing the response to treatment with other AML cohorts included in the study, KMT2A rearrangement was found to be significantly associated with a poorer response (p=0.0001), indicating that patients with KMT2A rearrangement have a higher likelihood of an unfavorable treatment outcome compared to patients with other AML genetic abnormalities.
Discussion
The analysis of genetic factors and gene rearrangement revealed that 66.1% of patients with Acute Myeloid Leukemia (AML) exhibited specific genetic abnormalities. On a global scale, cytogenetic abnormalities are observed in approximately 50–60% of newly diagnosed AML patients, and these abnormalities are characterized as non-random chromosomal rearrangements. These aberrations can be further categorized into three groups: AML with balanced translocations/inversions, AML with various cytogenetic abnormalities (such as unbalanced translocations, deletions, monosomies, and trisomies), and AML with complex karyotype (which involves the presence of at least three acquired chromosomal aberrations).6 Since in a prospective study, we reported the results of only the first category, the percentage of AML patients with genetic abnormalities tends to be high among Iraqi patients.
In the prospective study, the most common genetic transformation observed was the RUNX1::RUNX1T1 fusion, which was present in 16% of the AML patients. This finding is consistent with other studies that have reported a prevalence of 10–20% for the RUNX1::RUNX1T1 fusion in AML cases.7 However, it is noteworthy that a previous study conducted in Iraq by Al-Saidi et al reported a lower frequency of the RUNX1::RUNX1T1 fusion, with only 9% of AML patients exhibiting this genetic abnormality.8
The difference in the prevalence of the RUNX1::RUNX1T1 fusion between the current study and the study by Al-Saidi et al8 may be attributed to the different detection methods used. In the current study, PCR-based assays were employed for genetic analysis, which generally provide higher sensitivity and accuracy compared to fluorescence in situ hybridization (FISH) techniques. This difference in detection methods is particularly important in cases with low white blood cell (WBC) counts, where FISH may have reduced accuracy.
Also, a balanced translocation involving 11q23/KMT2A is among the most frequent chromosomal abnormalities in acute myeloid leukemia. In our study, approximately 20% of AML patients were found to have a translocation involving 11q23/KMT2A. This prevalence aligns with the findings of the study conducted by Issa et al, which also reported a similar frequency of approximately 20% for this translocation.9
In our study, which involved a substantial number of AML patients, we specifically examined cases with t(9;11) and other recurrent balanced rearrangements involving 11q23/KMT2A. Through our analysis, we identified both similarities and differences among AML patients with these specific translocations. Among the translocations, t(10;11) was found to be the most prevalent, followed by t(11;19), with frequencies ranging from 10% to 12%.10–12 These findings shed light on the genetic landscape of AML patients with translocations involving 11q23/KMT2A, providing insights into the prevalence and specific types of translocations observed. This knowledge contributes to a better understanding of the molecular basis of AML and may have implications for risk stratification and targeted therapeutic approaches in these patients.
In our study, we identified different molecular variants associated with Acute Promyelocytic Leukemia (APL). Specifically, we observed the presence of the PML-RARA fusion resulting from the t(15;17)(q24;q21) translocation in 6 (5.2%) patients, the PLZF-RARA fusion resulting from the t(11;17)(q23;q21) translocation in 3 (2.6%) patients, and the PLZF-RARA fusion resulting from the t(5;17)(q32;q21) translocation in 7 (6.1%) patients. These findings align with previous research by Liquori et al, who reported that the ZBTB16 (formerly PLZF)-RARA fusion is the most common molecular variant in APL.13
Interestingly, our prospective study revealed a higher prevalence of molecular variants compared to the PML-RARA fusion gene in APL cases. This finding contrasts with the results reported by Wen et al, who observed a very low percentage of APL patients with molecular variants.14 These discrepancies in findings may be attributed to differences in the detection techniques used and the relatively small sample size of APL cases in our current study. These findings underscore the heterogeneity of APL at the molecular level, with different fusion genes contributing to the disease. Understanding these molecular variants is crucial for accurate diagnosis and targeted treatment approaches in APL patients. Further research with larger sample sizes and comprehensive molecular profiling may provide deeper insights into the prevalence and clinical implications of these molecular variants in APL.
The chromosomal abnormality inv(16)(p13;q22) or t(16;16)(p13.1;q22) is found in approximately 5% to 10% of Acute Myeloid Leukemia (AML) cases.15 This genetic alteration is characterized by peripheral monocytosis and the infiltration of monoblasts in the bone marrow.15 In our prospective study, we observed that 5% of AML patients exhibited the CBFB::MYH11 fusion, which is indicative of inv(16)(p13;q22) or t(16;16)(p13.1;q22).
Upon performing the final risk stratification, we observed that 40% of the patients in our study were classified as having a favorable risk, primarily attributed to the presence of genetic abnormalities such as RUNX1-RUNX1T1 fusion, CBFB-MYH11 fusion, and NPM1 mutation. Intermediate risk was identified in 4.3% of the patients, mainly driven by the MLLT3-KMT2A fusion. Adverse risk stratification was assigned to 25.2% of the patients, predominantly due to the presence of genetic abnormalities such as DEK-NUP214 fusion, KMT2A rearrangement, and FLT-3 mutation. These findings align with the study conducted by Herold et al, which also reported similar percentages for the favorable and adverse risk groups.16 However, in their study, the intermediate risk group accounted for 20% of the patients. The discrepancy in percentages can be attributed to differences in sample size and the comprehensive molecular and genetic testing performed in their study, which included karyotyping analysis for all patients.
In our prospective study, we observed heterogeneous results among the different risk groups. However, there was a statistically significant increase in the number of deceased patients in the adverse risk group. This finding is consistent with previous studies that have also demonstrated a significant worsening of treatment response among patients classified as having adverse risk stratification.17,18 These results support the notion that risk stratification plays a crucial role in predicting treatment outcomes and overall prognosis in patients with acute myeloid leukemia. By identifying patients with adverse risk factors, healthcare providers can better tailor treatment approaches and interventions to improve patient outcomes.
The analysis of demographic data in relation to risk stratification revealed several significant associations. Advanced age, presence of comorbidities, and FAB classification (specifically M0 subtype) were found to be significantly associated with worsening of risk (p<0.05).19 These findings align with the understanding that AML is more prevalent in the elderly population, and prognosis tends to be poorer in older individuals.
Furthermore, a study by Prassek et al demonstrated that the spectrum and significance of driver gene mutations in older AML patients differ from those in younger patients. In the elderly population, various factors such as the presence of other medical conditions, lower functional ability, and preferences of both physicians and patients for less toxic treatment options often lead to the avoidance of intensive induction chemotherapy in favor of less aggressive therapies.20,21
Taken together, these findings highlight the complex interplay between demographic factors, disease characteristics, and treatment decisions in AML, particularly among elderly patients. The presence of comorbidities and advanced age can contribute to increased risk and influence treatment approaches, emphasizing the need for personalized management strategies in this population.
In the prospective analysis, the prevalence of CBF-AML was found to be higher at 21.7% among the study patients, surpassing the global trend of 12–15%.22 Encouragingly, a significant proportion of patients with CBF-AML achieved complete remission, with an observed rate of 84%. Only 4 patients (16%) experienced treatment failure, while no deaths were reported among those with CBF-AML during the study period. Comparing these outcomes with findings from other study cohorts, CBF-AML demonstrated a more favorable response. It is important to note that CBF-AML generally carries a better prognosis compared to other subtypes of AML. However, it is worth highlighting that despite the favorable prognosis, a subset of CBF-AML patients (approximately 30–40%) may still face the risk of relapse, potentially requiring allogeneic hematopoietic cell transplantation.23 Therefore, ongoing monitoring and appropriate management remain crucial for achieving long-term remission in CBF-AML cases.
Despite the utilization of All-trans retinoic acid (ATRA) as a treatment approach for APL, which has shown promising results in Iraq,24 notable differences in treatment response were observed in this study. Among patients with APL, all individuals with the PML-RARA fusion achieved complete remission CR. However, for patients with the PLZA-RARA fusion, two out of three patients died, and one experienced treatment failure (BP). In contrast, patients with the NPM-RARA fusion exhibited a CR rate of 71%. These findings highlight a significant disparity in treatment response, particularly with respect to patients harboring the PLZA-RARA fusion, who exhibited a poorer response compared to other rearrangements.
According to the 2017 ELN recommendations, patients diagnosed with t(9;11) are generally classified as intermediate-risk, regardless of their age or the presence of additional gene mutations. On the other hand, individuals with t(v;11)(v;q23) are categorized as adverse-risk.3 In the current study, patients with KMT2A rearrangement exhibited a poor response to treatment, with 65% experiencing treatment failure (BP), 30% of patients died, and only one patient achieving complete remission. Notably, there was no significant difference in treatment response observed among patients with KMT2A rearrangement. Compared to all other AML cohorts included in this study, KMT2A rearrangement was significantly associated with a poorer treatment response. Furthermore, our analysis revealed that this mutation was more commonly found in patients aged 60 years or older. These findings align with another recently published study, which proposed incorporating age into risk classification. Specifically, the presence of t(9;11) in younger patients may be considered intermediate-risk, whereas its presence in elderly individuals (above 60 years) would warrant adverse-risk stratification.25 These observations emphasize the importance of considering both genetic abnormalities and age when determining risk stratification and treatment approaches for patients with KMT2A rearrangement in order to optimize outcomes. Further research is needed to better understand the underlying mechanisms contributing to the poor response associated with KMT2A rearrangement and to develop targeted therapies for this specific subgroup of AML patients.
Conclusion
In summary, our prospective analysis of AML patients revealed a high prevalence of specific genetic abnormalities, exceeding the global average for such abnormalities in newly diagnosed AML cases. Additionally, we observed a higher occurrence of molecular variants in APL compared to the commonly studied PML-RARA fusion gene. The risk stratification based on genetic factors highlighted the significant associations of age, comorbidities, and FAB classification with risk classification. Notably, our study emphasized the importance of considering age when classifying risk, as the impact of certain genetic abnormalities may differ between younger and older patients. Further research with larger sample sizes is warranted to deepen our understanding of the molecular mechanisms and treatment responses in AML, particularly in subtypes such as APL and AML with KMT2A rearrangements.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Yu J, Li Y, Zhang D, Wan D, Jiang Z. Clinical implications of recurrent gene mutations in acute myeloid leukemia. Exp Hematol Oncol. 2020;9:4. doi:10.1186/s40164-020-00161-7
2. Hesham Moulod S, Ahmed AL-Rubaie H. April mRNA expression: does it predict the response to induction therapy in adult acute myeloid leukemia? Biochem Cell Arch. 2022;22:957–961.
3. Döhner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. J Am Soc Hematol. 2017;129(4):424–447.
4. National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) Acute Myeloid Leukemia Version 2.2022. Plymouth Meeting (PA): NCCN; 2022.
5. Tao S, Wang C, Chen Y, et al. Prognosis and outcome of patients with acute myeloid leukemia based on FLT3 ITD mutation with or without additional abnormal cytogenetics. Oncol Lett. 2019;18(6):6766–6774. doi:10.3892/ol.2019.11051
6. Pourrajab F, Zare-Khormizi MR, Hashemi AS, Hekmatimoghaddam S. Genetic characterization and risk stratification of acute myeloid leukemia. Cancer Manag Res. 2020;12:2231. doi:10.2147/CMAR.S242479
7. Gupta M, Mahapatra M, Saxena R. Cytogenetics’ impact on the prognosis of acute myeloid leukemia. J Lab Physicians. 2019;11(02):133–137. doi:10.4103/JLP.JLP_164_18
8. AL-Saidi DN, Hameed BM, Khaleel KJ. Detection of RUNX1-RUNX1T1 fusion gene in AML patients by FISH technique in Iraq. J Commun Dis. 2021;53(4):54–60.
9. Issa GC, Zarka J, Sasaki K, et al. Predictors of outcomes in adults with acute myeloid leukemia and KMT2A rearrangements. Blood Cancer J. 2021;11(9):1. doi:10.1038/s41408-021-00557-6
10. Cox MC, Panetta P, Lo-Coco F, et al. Chromosomal aberration of the 11q23 locus in acute leukemia and frequency of MLL gene translocation: results in 378 adult patients. Am J Clin Pathol. 2004;122(2):298–306. doi:10.1309/RX27R8GJQM330C22
11. Chen Y, Kantarjian H, Pierce S, et al. Prognostic significance of 11q23 aberrations in adult acute myeloid leukemia and the role of allogeneic stem cell transplantation. Leukemia. 2013;27(4):836–842. doi:10.1038/leu.2012.319
12. Bhatnagar B, Blachly JS, Kohlschmidt J, et al. Clinical features and gene-and microRNA-expression patterns in adult acute leukemia patients with t (11; 19)(q23; p13. 1) and t (11; 19)(q23; p13. 3). Leukemia. 2016;30(7):1586–1589. doi:10.1038/leu.2015.345
13. Liquori A, Ibañez M, Sargas C, Má S, Barragán E, Cervera J. Acute promyelocytic leukemia: a constellation of molecular events around a single PML-RARA fusion gene. Cancers. 2020;12(3):624. doi:10.3390/cancers12030624
14. Wen L, Xu Y, Yao L, et al. Clinical and molecular features of acute promyelocytic leukemia with variant retinoid acid receptor fusions. Haematologica. 2019;104(5):e195. doi:10.3324/haematol.2018.205369
15. Pulikkan JA, Castilla LH. Preleukemia and leukemia-initiating cell activity in inv (16) acute myeloid leukemia. Front Oncol. 2018;8:129. doi:10.3389/fonc.2018.00129
16. Herold T, Rothenberg-Thurley M, Grunwald VV, et al. Validation and refinement of the revised 2017 European LeukemiaNet genetic risk stratification of acute myeloid leukemia. Leukemia. 2020;34(12):3161–3172. doi:10.1038/s41375-020-0806-0
17. Acharya UH, Halpern AB, Wu QV, et al. Impact of region of diagnosis, ethnicity, age, and gender on survival in acute myeloid leukemia (AML). J Drug Assess. 2018;7:51–53. doi:10.1080/21556660.2018.1492925
18. Tallman MS, Wang ES, Altman JK, et al. Acute myeloid leukemia, Version 3.2019, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2019;17:721–749. doi:10.6004/jnccn.2019.0028
19. Prassek VV, Rothenberg-Thurley M, Sauerland MC, et al. Genetics of acute myeloid leukemia in the elderly: mutation spectrum and clinical impact in intensively treated patients aged 75 years or older. Haematologica. 2018;103:1853–1861. doi:10.3324/haematol.2018.191536
20. Rao AV. Fitness in the elderly: how to make decisions regarding acute myeloid leukemia induction. Hematology Am Soc Hematol Educ Program. 2016;2016(1):339–347.
21. AL-Khalissi KA. Experience with treatment of fifty eight Iraqi patients with Acute Myeloid Leukemia. JFacMedBagdad. 2014;55(4):290–295. doi:10.32007/jfacmedbagdad.554566
22. Borthakur G, Kantarjian H. Core binding factor acute myelogenous leukemia-2021 treatment algorithm. Blood Cancer J. 2021;11(6):1–5. doi:10.1038/s41408-021-00503-6
23. Ustun C, Morgan E, Moodie EE, et al. Core‐binding factor acute myeloid leukemia with t (8; 21): risk factors and a novel scoring system (I‐CBF it). Cancer Med. 2018;7(9):4447–4455. doi:10.1002/cam4.1733
24. Hussein AG, Ali MS, Hassan MS, Mansoor SS, Ameri AM AL, Muhsin RJ. Haematologic parameters in acute promyelocytic leukemia patients treated with ALL trans- retinoic acid. J Fac Med Bagdad. 2007;49(1):51–55.
25. Bill M, Mrózek K, Kohlschmidt J, et al. Mutational landscape and clinical outcome of patients with de novo acute myeloid leukemia and rearrangements involving 11q23/KMT2A. Proceed National Acad Sci. 2020;117(42):26340–26346. doi:10.1073/pnas.2014732117
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
