Back to Journals » ImmunoTargets and Therapy » Volume 12
The Methylation in B7-H4 and BTLA Genes are Associated with the Risk of Pulmonary Tuberculosis
Authors Cai XQ, Huang Q, Zhang TP
Received 8 August 2023
Accepted for publication 8 November 2023
Published 24 November 2023 Volume 2023:12 Pages 149—163
DOI https://doi.org/10.2147/ITT.S434403
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Michael Shurin
Xue-Qian Cai,1,* Qian Huang,2,* Tian-Ping Zhang3
1Department of Respiratory and Critical Care Medicine, Anhui Chest Hospital, Hefei, People’s Republic of China; 2Department of Public Health, Medical Department, Qinghai University, Xining, People’s Republic of China; 3The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, Anhui, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Tian-Ping Zhang, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, 17 Lujiang Road, Hefei, Anhui, 230001, People’s Republic of China, Email [email protected]
Background: The important roles of B7 homologous body 4 (B7-H4), B and T lymphocyte attenuator (BTLA) in patients with pulmonary tuberculosis (PTB) have been reported. This study aims to evaluate the association among B7-H4 and BTLA genes polymorphism, methylation and PTB susceptibility.
Methodology: Here, we assessed the possible relationship of 10 single nucleotide polymorphisms (SNPs) in B7-H4, BTLA genes with PTB susceptibility in a Chinese population (496 PTB patients and 502 controls) by SNPscan technique. Then, the B7-H4, BTLA genes methylation levels among 98 PTB patients and 97 controls were detected using MethylTarget technique.
Results: This study found no significant differences in allele and genotype frequencies of B7-H4 gene rs10754339, rs10801935, rs10923223, rs1937956, rs3738414, BTLA gene rs1982809, rs2971205, rs75368388, rs9288953 variants between PTB patients and controls. Haplotype analysis suggested that the lower frequencies of B7-H4 AATTG haplotype, BTLA GATT haplotype and the higher frequency of BTLA AGTC haplotype were found in PTB patients when compared with controls. We also found that the frequency of BTLA gene rs9288953 C allele was significantly increased in PTB patients with drug resistance. Moreover, the methylation levels of B7-H4 and BTLA genes in PTB patients were greater than that in controls, and rs10754339 variant in B7-H4 gene could affect its methylation level in PTB patients.
Conclusion: B7-H4, BTLA genes polymorphism might not affect PTB susceptibility, while the abnormal methylation levels of B7-H4, BTLA genes were associated with the genetic background of PTB.
Keywords: pulmonary tuberculosis, single nucleotide polymorphisms, DNA methylation, B7-H4, BTLA
Introduction
Tuberculosis (TB) is a common, serious infectious disease caused by Mycobacterium tuberculosis (M. tuberculosis) infection, which is still a major public health problem. Before the COVID-19 pandemic, TB was the leading cause of death from a single pathogen, and the most common type of this disease was pulmonary TB (PTB).1 The COVID-19 pandemic resulted in reduced access to TB diagnosis and treatment. There were approximately 10.6 million new cases of TB worldwide and about 0.78 million new cases of TB in China in 2021 according to the Global Tuberculosis Report 2022.2 Studies showed that about a quarter of the population was infected with M. tuberculosis globally, while less than 10% of those infected would develop active TB.3 This suggested that the pathogenesis of TB was influenced by multiple factors, including nutrition, immunity, genetic factors, smoke, alcohol, and HIV infection.4–6
Genome-wide association studies and twin studies had confirmed the important influence of host genetics on TB susceptibility.7,8 In addition, candidate gene association studies had found many important variants related to TB susceptibility, most of which were mainly located in immune-related genes, such as Toll-like receptor genes, vitamin D metabolic pathway genes and human leukocyte antigen genes.9–11 However, these studies only could explain part of the heritability of TB. Epigenetics also showed the important roles in TB development. For instance, abnormal DNA methylation, which was known as an important epigenetic mode, of several genes was considered to affect the development of TB.12
The PTB development was affected by the host’s immune function, and host control of M. tuberculosis infection required innate and adaptive immune responses. B- and T-lymphocyte attenuator (BTLA) and B7 homologous body 4 (B7-H4) were two newly discovered immune checkpoint inhibitors.13,14 BTLA was mainly expressed in dendritic cells (DC), T cells, B cells and some myeloid cells. The interaction between BTLA and its ligand herpesvirus entry mediator (HVEM) could inhibit the proliferation and activation of T cells, as well as the expression of a variety of cytokines in T cells.15 The function of BTLA in PTB had been investigated, and BTLA exerted immune memory on αβT cells in active PTB.16 B7-H4 was inductively expressed by immune cells such as T cells, B cells, DC cells and macrophages. The increased expression of B7-H4 in DC might be associated with the increase of Tregs, and could influence the differentiation of naive T cells, thereby affecting the identification of M. tuberculosis by the immune system and inducing anti-TB specific immune responses.17 BTLA and B7-H4 expression levels were abnormally increased in CD11c+ antigen presenting cells from peripheral blood among PTB patients, suggesting that these factors were closely involved in the pathogenesis of PTB.18
Previous studies revealed that genetic variation in B7-H4, BTLA was related the susceptibility to many diseases, including autoimmune thyroid diseases,19 ankylosing spondylitis (AS),20 breast cancer.21 Nevertheless, the genetic effect of B7-H4, BTLA genes on the risk of PTB had not yet been explored. Therefore, we conducted this study to investigate the association among several single nucleotide polymorphisms (SNPs) in B7-H4, BTLA genes, as well as their methylation level, and PTB susceptibility in the Chinese Han population.
Materials and Methods
PTB Patients and Controls
We continuously collected the patients with PTB from Anhui Chest Hospital, and all patients were diagnosed by senior clinicians according to the diagnosis for PTB (WS 288–2017) and the classification of TB (WS 196–2017) from the Health Industry Standard of the People’s Republic of China. Exclusion criteria of PTB patients were: HIV-positive, other infectious diseases, autoimmune diseases and cancer. In this study, we selected healthy volunteers from the local health examination center as control group, and all of whom had no history of infectious diseases, pulmonary diseases, bacterial or viral infections.
The study was conducted in compliance with the provisions of the Declaration of Helsinki and approved by the Ethics Committee of Anhui Chest Hospital (K2023-011). Written informed consent was obtained from all participants, peripheral blood (3–5 mL), and clinical data were collected. Genomic DNA was extracted from peripheral blood using the Flexi Gene-DNA Kit (Qiagen, Valencia, CA). The required clinical data included fever, drug resistance, drug-induced liver injury (DILI), pulmonary infection, leukopenia, sputum smear, etc.
SNP Selection and Genotyping
A total of ten SNPs, including B7-H4 gene rs10754339, rs10801935, rs10923223, rs1937956, rs3738414 and BTLA gene rs9288952, rs1982809, rs2971205, rs75368388, rs9288953 were genotyped by SNPscan technique, with technical support from the Center for Genetic and Genomic Analysis, Genesky Biotechnologies Inc., Shanghai. Only participants with 100% genotyping success for the selected SNPs were included in the final analysis.
The selection of these SNPs was mainly based on the following principles of screening tag SNPs. The tag SNPs of B7-H4 and BTLA genes was selected using genetic data from Ensembl genome browser 85 and CHBS_1000g with Haploview 4.0 software (Cambridge, MA, USA) based on following strategy: (1) minor allele frequency ≥0.05; (2) an r2 cutoff of 0.8 for linkage disequilibrium; (3) the flanking 2.0 kb regions of each gene.
Methylation Detecting
In this study, DNA samples were first treated with bisulfite, and then used for targeted region methylation sequencing to the promoter regions in B7-H4 and BTLA genes. The CpG islands of the target fragments in the promoter regions of these two genes were sequenced using the MethylTarget technique with the support of the Genesky Biotechnology (Shanghai) Center for Genetic and Genome Analysis. Only one specific target fragment was respectively detected on B7-H4 and BTLA genes, and the primer sequence of these fragments were shown in Table S1. The methylation level of every specific target fragment could be got through calculating the mean methylation level of all CpG sites on the fragment.
Statistical Analysis
We used the chi-squared test to assess whether the genotype distribution of 10 SNPs in B7-H4 and BTLA genes in the control group achieved Hardy–Weinberg Equilibrium (HWE). The distributions of allele and genotype between PTB patients and controls were compared by logistic regression model, and odds ratios (OR) with 95% confidence intervals (CI) was calculated. The association of B7-H4 and BTLA genes variation with PTB susceptibility was analyzed under two genetic models (dominant model and recessive model), and haplotype analysis was conducted using SHEsis software.22 The methylation levels of B7-H4 and BTLA genes were expressed by median (P25, P75). The differences of B7-H4 and BTLA genes methylation levels between different groups were analyzed by Mann–Whitney U-test and Kruskal–Wallis H-test. All statistical analyses were calculated with SPSS 23.0, and a P-value <0.05 was regarded as statistically significant. Bonferroni correction was used for multiple testing in SNP analysis, and P<0.0055 (0.05/9) was considered as statistically significant.
Results
Comparison of B7-H4 and BTLA Genes Polymorphisms Among PTB Patients and Controls
Ten SNPs in B7-H4 and BTLA genes was successfully genotyped in 496 patients with PTB and 502 controls. The differences regarding age (an average age of patients: 44.73 ± 18.04 years, an average age of controls: 44.42 ± 3.50 years) and gender (patients: 183 female/313 male; controls: 201 female/301 male) distribution between PTB patients and controls showed no significant. HWE test was performed for the genotype distribution of all SNPs in controls, and the results showed that nine SNPs exhibited HWE except BTLA gene rs9288952.
Our results revealed that GA genotype frequency of rs3738414 variant in B7-H4 gene was increased in PTB patients when compared to controls (P=0.030 after adjustment of gender and age), while the difference was not statistically significant after Bonferroni correction (P>0.0055) (Table 1). Similarly, there was no statistical association between the allele, genotype frequencies of other SNPs (rs10754339, rs1937956, rs10923223, rs10801935) in B7-H4 gene and PTB susceptibility. For BTLA gene, the frequencies of AA genotype in rs1982809 variant, GG genotype in rs2971205 variant, C allele in rs9288953 variant seemed to be increased in PTB patients than that in controls (P=0.039, P=0.018, P=0.045 after adjustment of gender and age), but no statistically significant association was found after Bonferroni correction (P>0.0055) (Table 1). The association among B7-H4, BTLA genes polymorphism and PTB susceptibility was also assessed under dominant model, recessive mode, and no association reached statistical significance after Bonferroni correction (P>0.0055).
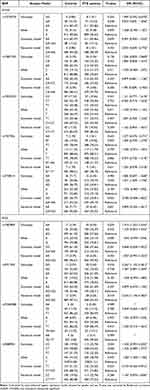 |
Table 1 Association of B7-H4, BTLA Genes Polymorphisms with PTB Susceptibility |
We constructed the haplotypes of B7-H4 and BTLA genes by SHEsis software, and only the haplotypes with a frequency more than 3% were included in Table 2. The results of haplotype analysis showed that when compared with controls, AATTG haplotype of B7-H4 gene and GATT haplotype of BTLA gene had significantly decreased frequencies (P=0.033, P=0.045) in PTB patients, while AGTC haplotype of BTLA gene had an increased frequency (P=0.002). Other haplotypes of B7-H4 and BTLA genes did not exhibit any significant association with PTB susceptibility.
 |
Table 2 Haplotype Analysis of B7-H4, BTLA Genes in PTB Patients |
Association Between B7-H4, BTLA Genes Polymorphisms and Several Clinical Manifestations in PTB Patients
We respectively divided PTB patients into two groups based on whether the patients had specific clinical features or not, such as fever, drug resistance, DILI, pulmonary infection, hypoproteinemia, sputum smear-positive. Then, we analyzed the association between the allele and genotype frequencies of all SNPs in B7-H4, BTLA genes and above clinical manifestations in PTB patients. The results demonstrated that the frequency of BTLA gene rs9288953 C allele was significantly increased in PTB patients with drug resistance in comparison with the patients without drug resistance (P=0.035) (Table 3). However, other SNPs in B7-H4 and BTLA genes were not associated with clinical manifestations of PTB.
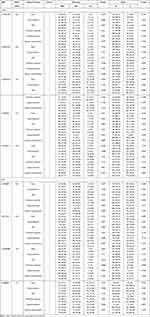 |
Table 3 Relationship Between B7-H4, BTLA Genes Polymorphisms and the Clinical Manifestations in PTB Patients |
Comparison of B7-H4, BTLA Genes Methylation Levels in PTB Patients and Controls
The methylation levels of B7-H4, BTLA genes were examined in 98 PTB patients and 97 controls. No difference in the distribution of age (an average age of patients: 43.52 ± 17.59 years, an average age of controls: 43.45 ± 4.49 years) or gender (patients: 45 female/53 male; controls: 46 female/51 male) between PTB patients and controls was found. In this study, only one specific target fragment was detected in B7-H4 and BTLA genes, respectively. Therefore, the methylation level of every target fragment was used to represent the methylation level of B7-H4 and BTLA genes in this study.
In PTB patients, the methylation levels of B7-H4, BTLA genes were 0,2966(0.2938-0.3003), 0.7735 (0.7171–0.8433). In the control group, the methylation level of B7-H4, BTLA genes were 0.2941(0.2914-0.7925), 0.7456 (0.7149–0.7925). As shown in Figures 1 and 2, we found that the methylation levels of B7-H4, BTLA genes in PTB patients were both higher than that in controls (P=0.001, P=0.005). We also analyzed the association among B7-H4, BTLA genes methylation levels and some clinical manifestations in PTB patients. This study did not obtain any significant association between B7-H4, BTLA genes methylation levels and the clinical manifestations, including fever, drug resistance, DILI, pulmonary infection, hypoproteinemia, sputum smear-positive (Table 4).
 |
Figure 1 The methylation levels of B7-H4 genes in PTB patients and controls. |
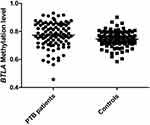 |
Figure 2 The methylation levels of BTLA genes in PTB patients and controls. |
 |
Table 4 Association Between B7-H4, BTLA Methylation Levels and Clinical Characteristics of PTB Patients |
Association Between B7-H4, BTLA Genes Polymorphisms and Their Methylation Levels
This study conducted an association analysis to assessed whether the genotype of each SNP of B7-H4, BTLA genes affected their methylation levels in 98 PTB patients (Table 5). Our results demonstrated that rs10754339 variant in B7-H4 gene could affect its methylation level. In addition, no statistically significant association was found among BTLA gene polymorphism and its methylation level.
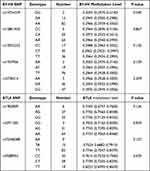 |
Table 5 Association between B7-H4, BTLA genes genotypes on their methylation levels |
Discussion
As a complex infectious disease, genetic factors played an important role in the pathogenesis of PTB. Further elucidation of host genetic differences was an important way to understand the pathogenesis of PTB and formulate appropriate treatment, prognosis strategies. Recently, some studies had extensively studied the regulatory role of checkpoint inhibitory molecules BTLA and B7-H4 in a variety of immune cells, and revealed their influence on the pathogenesis of PTB.16,23 The present study was the first to explore the potential association among BTLA, B7-H4 genes variation and PTB susceptibility from the perspective of gene polymorphisms and epigenetics, and provided several previously undescribed findings.
B7-H4, also known as B7S1 and B7x, was the newest member of the B7 family, and could modulate antigen-specific immune responses by inhibiting T cell activation and the production of cytokines.24 One previous research demonstrated that B7-H4-expressing macrophages was significantly increased in the peripheral blood from lung cancer patients when compared to TB patients and healthy controls.25 In addition, the high expression of B7-H4 was not only positively related to tumor progression, but also involved in tumor immune escape. Chen et al found that mature activated DCs expressed CD83 and several negative immune molecules, such as B7-H4, BTLA,25 and an obvious proportion increase regarding the co-expression of BTLA and B7-H4 was found in mDC from TB patients.17 Moreover, this study also implied that the high expression of B7-H4 and BTLA might play a negative regulatory influence of the mDCs on anti-TB immunity through promoting the expression of CD83 and HLA-DR. Some studies were also conducted the potential connection and the polymorphism in B7-H4 gene and susceptibility to human disease. Zhang et al performed an association study of rs10754339, rs10801935, rs3738414 variants in B7-H4 gene in breast cancer, and found that these SNPs were connected with the risk of breast cancer.26 The results by Chen et al indicated that two polymorphisms (rs10801935, rs3738414) in B7-H4 gene were significantly associated with AS susceptibility, and TAG haplotype was a risk factor for AS by haplotype analysis.20 Therefore, we speculated that B7-H4 gene polymorphism might also be related to the susceptibility to PTB, and analyzed the association between five SNPs (rs10754339, rs10801935, rs10923223, rs1937956, rs3738414) in B7-H4 gene with PTB risk. The rs3738414 GA genotype was found to increase susceptibility to PTB, but this association was not statistically significant after Bonferroni correction. Furthermore, AATTG haplotype in the B7-H4 gene might be associated with an increased risk of PTB. This suggested that B7-H4 gene variation might be involved in the pathogenesis of PTB, but further verification was needed.
As a newly discovered inhibitory receptor, BTLA exerted important roles in suppressing the overreactive immune response and could play immunosuppressive effects on the host by activating B cells and T cells.27 Recent evidences described the important function of BTLA in many diseases, including inflammatory disorder, autoimmune diseases and PTB.23,28 Previous studies regarding the roles of BTLA on PTB mainly focused on its expression level in several specific cell.18,23 Wang et al revealed the effect of BTLA in CD11c antigen presenting cells (APCs) in active PTB patients, and found that BTLA-expressing CD11c APCs showed a stronger ability to stimulate T cell proliferation.18 Another study demonstrated that the expression of CD11c+ BTLA was positively related to CRP, which might be correlated with inflammatory changes in PTB patients.29 The present study mainly focused on studying the association of four SNPs (rs1982809, rs2971205, rs75368388, rs9288953) in BTLA gene with PTB risk, while no significant association was found between these SNPs and the risk of PTB. Contrary to our study, previous studies displayed significant correlation between BTLA gene polymorphisms and cancer, autoimmune diseases. Andrzejczak et al found that rs1982809 variant was significantly related to non-small cell lung cancer risk.30 Karabon et al reported that rs1982809, rs9288953 variants would affect the susceptibility to chronic lymphocytic leukemia.31 It was worth noting that rs9288953 variant was found to be significantly associated with the development of drug resistance among PTB patients. Therefore, BTLA gene polymorphism could be used as a potential index to determine the clinical classification of PTB patients.
In addition to DNA sequences, important genetic information was also present in epigenetic variations.32 Gene polymorphism and epigenetic mechanisms might interact and together influenced the biological function and disease progression.33 Therefore, the regulatory role of abnormal DNA methylation, which was considered as a common epigenetic trait, of certain key genes in human diseases had been reported.34 DNA methylation had become an irreplaceable way to explore the pathogenesis of PTB. The results of Wang et al showed that abnormal methylation status of several key genes in the vitamin D metabolic pathway was closely connected with the risk and prognosis of PTB.12 Considering the potential significance of B7-H4 and BTLA genes polymorphisms in PTB development, this study further analyzed the relationship among their methylation levels and PTB risk. Here, the present results demonstrated that the methylation levels of B7-H4 and BTLA genes were significantly increased in PTB patients. Interestingly, we found that B7-H4 methylation level was affected rs10754339 variants among PTB patients. This implied that abnormal methylation levels of B7-H4 and BTLA genes were involved in the pathogenesis of PTB, and there might be an interaction between B7-H4 gene polymorphism and its methylation in PTB patients. The above findings provided important clues to reveal the occurrence of PTB, while relevant results still needed to be further verified.
Some limitations of this study should be noted. First, our study only screened several tagSNPs in B7-H4 and BTLA genes, and some SNPs with important functions might be missed. Second, we compared methylation levels of B7-H4 and BTLA genes between PTB patients and controls in a cross-sectional study, and did not analyze their changes levels over the course of PTB development. Third, our study did not exclude the possible influence of environmental factors and ethnic background on the results.
In summary, B7-H4, BTLA genes polymorphism might not be affected PTB susceptibility in the Chinese population, while the abnormal methylation levels of B7-H4, BTLA genes were associated with PTB risk. These results firstly revealed the roles of B7-H4, BTLA genes methylation in the development of PTB, which was important for PTB control and prevention. Moreover, the further studies regarding the mechanism of B7-H4, BTLA genes variation on PTB development was still warranted in future.
Ethics Approval and Consent to Participate
This study was approved by the Ethical Committee of Anhui Chest Hospital (K2023-011). All the study subjects provided informed consent to participate in this study.
Data Sharing Statement
The data generated and analyzed by this study are available from the corresponding author on reasonable request.
Funding
There is no funding to report.
Disclosure
Xue-Qian Cai and Qian Huang are co-first authors for this study. The authors report no conflicts of interest in this work.
References
1. Bai H, Song M, Lei S, et al. Genome-wide association study of tuberculosis in the western Chinese Han and Tibetan population. Med Comm. 2023;4(2):e250.
2. World Health Organization. Global tuberculosis report 2022. Available from: https://www.who.int/teams/global-tuberculosis-programme/TB-reports/global-tuberculosis-report-2022.
3. Aravindan PP. Host genetics and tuberculosis: theory of genetic polymorphism and tuberculosis. Lung India. 2019;36(3):244–252. doi:10.4103/lungindia.lungindia_146_15
4. Dye C, Lonnroth K, Jaramillo E, Williams BG, Raviglione M. Trends in tuberculosis incidence and their determinants in 134 countries. Bull World Health Organ. 2009;87(9):683–691. doi:10.2471/BLT.08.058453
5. Gupta M, Srikrishna G, Klein SL, Bishai WR. Genetic and hormonal mechanisms underlying sex-specific immune responses in tuberculosis. Trends Immunol. 2022;43(8):640–656. doi:10.1016/j.it.2022.06.004
6. de Lima D S, Morishi Ogusku M, Santos M PD, et al. Alleles of HLA-DRB1*04 associated with pulmonary tuberculosis in amazon Brazilian population. PLoS One. 2016;11(2):e0147543. doi:10.1371/journal.pone.0147543
7. Möller M, Hoal EG. Current findings, challenges and novel approaches in human genetic susceptibility to tuberculosis. Tuberculo. 2010;90(2):71–83. doi:10.1016/j.tube.2010.02.002
8. Zheng R, Li Z, He F, et al. Genome-wide association study identifies two risk loci for tuberculosis in Han Chinese. Nat Commun. 2018;9(1):4072. doi:10.1038/s41467-018-06539-w
9. Mhmoud NA. Association of toll-like receptors 1, 2, 4, 6, 8, 9 and 10 genes polymorphisms and susceptibility to pulmonary tuberculosis in Sudanese patients. Immunotargets Ther. 2023;Volume 12(12):47–75. doi:10.2147/ITT.S404915
10. Zhang TP, Chen SS, Zhang GY, Shi SJ, Wei L, Li HM. Association of vitamin D pathway genes polymorphisms with pulmonary tuberculosis susceptibility in a Chinese population. Genes Nutr. 2021;16(1):6. doi:10.1186/s12263-021-00687-3
11. Wang X, Cao X, Zhang W, et al. Association of human leukocyte antigens-DQB2/DPA1/DPB1 polymorphism and pulmonary tuberculosis in the Chinese Uygur population. Mol Genet Genomic Med. 2019;7:3.
12. Wang M, Kong W, He B, et al. Vitamin D and the promoter methylation of its metabolic pathway genes in association with the risk and prognosis of tuberculosis. Clin Epigenetics. 2018;10(1):118. doi:10.1186/s13148-018-0552-6
13. Poh SL, Linn YC. Immune checkpoint inhibitors enhance cytotoxicity of cytokine-induced killer cells against human myeloid leukaemic blasts. Cancer Immunol, Immunotherapy. 2016;65(5):525–536. doi:10.1007/s00262-016-1815-8
14. Shrestha R, Prithviraj P, Anaka M, et al. monitoring immune checkpoint regulators as predictive biomarkers in hepatocellular carcinoma. Front Oncol. 2018;8:269. doi:10.3389/fonc.2018.00269
15. Sedy JR, Gavrieli M, Potter KG, et al. B and T lymphocyte attenuator regulates T cell activation through interaction with herpesvirus entry mediator. Nat Immunol. 2005;6(1):90–98. doi:10.1038/ni1144
16. Zeng JC, Lin DZ, Yi LL, et al. BTLA exhibits immune memory for αβ T cells in patients with active pulmonary tuberculosis. Am J Transl Res. 2014;6(5):494–506.
17. Cai X, Ge N, Rong R, Lu Y, Zhang J, Xu J. High expression of BTLA and B7-H4 on the surface of myeloid dendritic cells has a negative regulatory effect on their anti-tuberculosis immunity activity in pleural tuberculosis patients. Tuberculosis. 2019;119:101877. doi:10.1016/j.tube.2019.101877
18. Wang WD, Gao YC, Lu YB, et al. BTLA-expressing CD11c antigen presenting cells in patients with active tuberculosis exhibit low capacity to stimulate T cell proliferation. Cell Immunol. 2017;311:28–35. doi:10.1016/j.cellimm.2016.09.015
19. Ozaki H, Inoue N, Iwatani Y, et al. Association of B7H3 and B7H4 gene polymorphisms and protein expression with the development and prognosis of autoimmune thyroid diseases. Clin Endocrinol. 2023;99(1):103–112. doi:10.1111/cen.14923
20. Chen Y, Yang H, Xu S, et al. Association analysis of B7-H3 and B7-H4 gene single nucleotide polymorphisms in susceptibility to ankylosing spondylitis in eastern Chinese Han population. Int J Immunogenet. 2021;48(6):500–509. doi:10.1111/iji.12559
21. Zhao RP, Li Z, Li C, et al. A genetic variant of the BTLA gene is related to increased risk and clinical manifestations of breast cancer in Chinese women. Clin Breast Cancer. 2021;21(5):e512–e517. doi:10.1016/j.clbc.2020.12.009
22. Li Z, Zhang Z, He Z, et al. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis. Cell Res. 2009;19(4):519–523. doi:10.1038/cr.2009.33
23. Shen X, Zhang J, Tang P, et al. Expression and clinical significance of B and T lymphocyte attenuator on CD4+ and CD8+ T cells from patients with pulmonary tuberculosis. Indian J Pathol Microbiol. 2019;62(2):232–238. doi:10.4103/IJPM.IJPM_727_17
24. Kryczek I, Wei S, Zou L, et al. Cutting edge: induction of B7-H4 on APCs through IL-10: novel suppressive mode for regulatory T cells. J Immunol. 2006;177(1):40–44. doi:10.4049/jimmunol.177.1.40
25. Chen C, Zhu YB, Shen Y, Zhu YH, Zhang XG, Huang JA. Increase of circulating B7-H4-expressing CD68+ macrophage correlated with clinical stage of lung carcinomas. J Immunother. 2012;35(4):354–358. doi:10.1097/CJI.0b013e31824212c4
26. Zhang J, Zhang M, Jiang W, et al. B7-H4 gene polymorphisms are associated with sporadic breast cancer in a Chinese Han population. BMC Cancer. 2009;9(1):394. doi:10.1186/1471-2407-9-394
27. Ning Z, Liu K, Xiong H. Roles of BTLA in Immunity and Immune Disorders. Front Immunol. 2021;12:654960. doi:10.3389/fimmu.2021.654960
28. Wojciechowicz K, Spodzieja M, Lisowska KA, Wardowska A. The role of the BTLA-HVEM complex in the pathogenesis of autoimmune diseases. Cell Immunol. 2022;376:104532. doi:10.1016/j.cellimm.2022.104532
29. Xu F, Bian K, Wang S, et al. B and T lymphocyte attenuator as a C-reactive protein and IgA associated auxiliary diagnostic marker for pulmonary tuberculosis: a case-control study. Ann Transl Med. 2022;10(24):1370. doi:10.21037/atm-22-6060
30. Andrzejczak A, Partyka A, Wiśniewski A, et al. The association of BTLA gene polymorphisms with non-small lung cancer risk in smokers and never-smokers. Front Immunol. 2023;13:1006639. doi:10.3389/fimmu.2022.1006639
31. Karabon L, Partyka A, Jasek M, et al. Intragenic Variations in BTLA Gene Influence mRNA Expression of BTLA Gene in chronic lymphocytic leukemia patients and confer susceptibility to chronic lymphocytic leukemia. Arch Immunol Ther Exp. 2016;64(Suppl 1):137–145. doi:10.1007/s00005-016-0430-x
32. Shnorhavorian M, Schwartz SM, Stansfeld B, Sadler-Riggleman I, Beck D, Skinner MK. Differential DNA methylation regions in adult human sperm following adolescent chemotherapy: potential for epigenetic inheritance. PLoS One. 2017;12(2):e0170085. doi:10.1371/journal.pone.0170085
33. Olsson AH, Volkov P, Bacos K, et al. Genome-wide associations between genetic and epigenetic variation influence mRNA expression and insulin secretion in human pancreatic islets. PLoS Genet. 2014;10(11):e1004735. doi:10.1371/journal.pgen.1004735
34. Farsetti A, Illi B, Gaetano C. How epigenetics impacts on human diseases. Eur J Intern Med. 2023;114:15–22. doi:10.1016/j.ejim.2023.05.036
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
