Back to Journals » Pathology and Laboratory Medicine International » Volume 15
Whole Slide Imaging Integration with Lab Information Systems, a Study of the Requirements, Processes and Procedures Enabling a Reporting-Based Workflow
Authors Alchami FS, Iqbal Z, Björkhammer CN, Saeed MO, Ramakrishnan R, Clelland C, Ahmad F, Charles A
Received 7 September 2022
Accepted for publication 28 December 2022
Published 17 January 2023 Volume 2023:15 Pages 1—11
DOI https://doi.org/10.2147/PLMI.S388981
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Paul Zhang
Fouad S Alchami,1,2 Zafar Iqbal,3 Carl Niclas Björkhammer,1 Mohammed O Saeed,3 Ramachandran Ramakrishnan,3 Colin Clelland,1 Fareed Ahmad,3 Adrian Charles1
1Department of Pathology, Sidra Medicine, Doha, Qatar; 2Department of Pathology and Laboratory Medicine, Weill Cornell Medicine, Qatar, Doha, Qatar; 3Imaging Information Systems, Sidra Medicine, Doha, Qatar
Correspondence: Fouad S Alchami, Department of Pathology, Sidra Medicine, Doha, 26999, Qatar, Tel +974 40033034, Email [email protected]
Background: Significant advances have been achieved in the clinical implementation of digital pathology (DP) whole slide imaging (WSI). However, the workflow of the reporting process and the full integration of this technology into the lab and hospital information system have not been sufficiently addressed. There is not sufficient vendor and industry consideration for this in the development and implementation process.
Methods: Exploring the requirements of direct integration of WSI with the lab and hospital information systems, demonstrating the workflow, challenges and best practices. Mapping and refining the process of movement of the digital slides. The anatomical pathology team, in collaboration with information technology and the vendor, establish the required scripts in order to enable direct integration, therefore allowing the laboratory a highly automated system of case, slide, and report movement.
Results: Direct integration was achieved fulfilling the required goals specified by the workflow mapping. An automated case movement process was established in order to enable the pathologist a work management process from the digital platform. The integration errors and challenges were audited and reported.
Conclusion: WSI digital pathology integration is achievable; however, it requires significant resource allocation adding to the technology acquirement.
Keywords: digital pathology, whole slide imaging, lab information systems, integration
Background
Digital pathology is widely accepted and validated for clinical use and is already in practice in multiple countries.1,2 Philips IntelliSite Pathology Solution (PIPS), the system in use and clinically live at Sidra Medicine, is the first whole slide imaging (WSI) system approved by the US Department of Health and Human Services Food and Drug Administration (FDA).3,4 Other WSI providers, such as Leica Biosystems, 3DHISTECH, and Sectra etc., have already been approved or are undergoing the approval process. The image management systems (IMS) available vary widely and currently do not offer any preexisting integration with known lab information systems (LISs) or hospital information systems (HISs). This does not take into consideration the complexity of the anatomical pathology reporting process requiring viewing of clinical request and notes, grossing, requesting extra stains and composing a report.5 Purchasing an FDA-approved WSI system does not include a pre-fabricated and tested direct integration with the major lab information system (LIS). Institutions currently using a WSI for primary reporting have largely resorted to local LIS-driven integration; such integration is achieved with locally engineered LIS and involves a costly and lengthy process if applied for commercial LIS. It also requires a mutual agreement between the providers of the LIS and IMS. Other institutions required the purchase of additional software to enable integration. To date, there is no commercial software available for direct integration to enable an anatomical pathology IMS-driven workflow for the reporting process through Cerner Pathnet. Also, there is no agreement on an industry-wide process for software reporting workflow for anatomical pathologists, an important step in an evolving industry. There have recently been some recommendations to address the integration requirements.6
In the implementation of WSI, the integration workflow of the pre-reporting process, i.e. viewing the slides, and its importance for ensuring quality is widely studied; however, addressing the reporting workflow for the pathologist has not been sufficiently considered.7,8 Briefly, the reporting process of the WSI could be launched either using an IMS-driven workflow or an LIS-driven workflow. The current workflow for reporting in institutions using WSI is mainly based on an LIS-driven workflow. In such a workflow the pathologist launches the images through a link in LIS or by logging separately and launching these applications independently, rendering the IMS an image projector rather than a complete technology solution.9
An IMS-driven workflow enables the pathologist to view and manage the case using the IMS. Such a workflow is widely used for radiology reporting, known as a picture archiving and communication system (PACS) driven workflow. It also is very similar to pathologists using the stack of slides and hard copy of the requests to prioritize and triage the workflow. An IMS-driven workflow would unlock the networking potential of WSI digital pathology amongst institutions harboring different LISs, a factor used in the justification and business case planning for the WSI adaptation.10,11 Unlocking the networking potential, irrespective of different institutional LISs, enables easier integration of reporting with image analysis applications,12 and is a key element in addressing the current and future workforce challenges in pathology.13 In radiology (PACS) implementation, the significant differences in vendor-specific image design and data communication between individual PACS components and radiology information systems (RISs) was a major challenge.14,15 PACS has now evolved allowing the majority of PACS to report from the application and publishing the report in the electronic medical record (EMR) without the need to access the EMR independently, through the use of well-defined communicating messages expected from the providers throughout the industry. Having the LIS integrated with the HIS adds another layer of complexity, but it is helpful for the pathologist to access the EMR, radiology and other pathology reports and images.
There are wide differences in the structures of the LISs and EMR systems used worldwide. These also vary between in-house developed systems and commercially available systems. This coupled with the variations in the design of the IMSs creates the greatest difficulty in adequate integration of the reporting process. This deficiency is leading to the use of multiple systems to do the one task of reporting.16 The LIS in use in our institution is Cerner Millennium™, one of the most widely used systems, with 27,500 client facilities in over 35 countries, which integrates the HIS and the LIS components.17
The objective is to establish the highest level of integration between IMS and LIS in order to establish an IMS-driven reporting workflow using WSI digital pathology, whilst ensuring quality. In doing this it highlights an industry-wide discussion about the technological requirements for such integrated workflows across different providers.
Methods
Institution, Hardware and Software
Sidra Medicine is a 400-bed tertiary women and children’s hospital in the State of Qatar, affiliated with Weill Cornell Medical College in Qatar. Inaugurated in 2016, its vision is to apply up to date evidence-based medicine using latest technology to its patients, including digital pathology. The anatomical pathology department is the main pediatric pathology provider for the State of Qatar, serving a population of around 2.7 million.
Sidra Medicine uses Cerner Millennium (Cerner Corp, Kansas City, US), recently upgraded to Cerner Code Upgrade 2018.08, as its electronic health care record management system. The anatomical pathology division utilizes the Cerner AP PathNet application, for case management of anatomical pathology cases. The AP PathNet add-on application, Advanced Barcode & Tracking (AB&T) is used for the tracking of all inventory, including uniquely identifiable specimens, blocks and slides, minimizing the risk of mis-identification throughout the process.
All routinely cut sections are stained with Hematoxylin & Eosin using a Leica ST5020 (Leica Microsystems, UK). Special stains are performed using a special stains instrument (Dako Artisan, Dako) and immunohistochemistry (IHC) is performed via an IHC platform (BOND III, Leica Microsystems). Canon EOS 5D digital (Canon, USA), is used for macroscopic imaging.
All generated slides are scanned using one of the two high volume throughput Philips ultra-fast scanners (UFSs) or the more versatile, single stage, low volume, ultra versatile scanner (UVS) (Philips, The Netherlands). Philips IntelliSite Pathology Solution (version 3.2) and its image management system (IMS) (version 3.3.1) is used as the sole WSI viewing platform.
Integration and Imaging Team
The project is led by an attending physician (FA) in anatomical pathology and a senior technologist (CB) to generate, examine, and audit the experimentation and validation.
Imaging support team: it was identified during the early stages of this project that imaging specialist personnel with LIS, RIS, and PACS experience will provide a fresh and evolved outlook regarding the potential level of integration WSI can achieve, taking the digitalized radiology story into consideration. This was also required to oversee the technical installation and configuration to implement this full system across the two data centers at Sidra.
The hospital’s dedicated in-house Cerner experts with extensive experience in Health Level Seven (HL7) messaging were essential. Philips contractually provided the required support in the integration and applying of the required customization and changes in the IMS.
Determining Levels of Integration and Developing a Functional Integration View
The planning for the integration view and workability of the reporting process for an IMS-driven workflow was constrained to the functionality of the IMS as specified by the vendor. The IMS therefore did not allow for additional tab insertion. A tab for report linking with the LIS is an example of much-needed integration.
Integration and validation scripts: the integration and validation scripts were based upon slide generated functions in PathNet (the AP LIS component of Cerner Millennium). The validation and testing took into consideration the various timelines, slide types, cancellation and recreation of slide requesting processes. This was done using test patient in Cerner Millennium, followed by the request for multiple tissue pathology requests using the Cerner PowerChart7 application simulating the requesting process by the clinical team. The relevant slides for scanning were generated through following a mock specimen from grossing, embedding, cutting and staining. The barcode label, the main linking identifier between the IMS and HL7 messages, was generated through Cerner PathNet. Integration issues were reported to the integration team via email and data collected. Prior to clinical validation, using College of American Pathologists (CAP) standards, a period of six months enabled ironing out of unforeseen issues by scanning surgical pathology slides generated and auditing system errors and problems in functionality.
Results
Pre-Scanning Requirement Satisfaction for Integration
The slide barcode generated by Cerner was used as the slide identifier linking various steps and tasks to the HL7 messages to be used in integrating the IMS. This was considered, planned for and achieved during the setup of the laboratory. Briefly, Cerner PathNet and Advanced Barcode & Tracking (AB&T) enabled traceability of slide requests, blocks and slides throughout the grossing and cutting stages. We achieved full barcode integration between Cerner and Leica ST5020 (Leica Microsystems, UK), Dako Artisan (Dako, Denmark) and BOND III (Leica Microsystems, UK) for Hematoxylin & Eosin stained slides and, special stains and immunohistochemistry (IHC), respectively. The latter two took significant time and effort. Macroscopic images taken using Canon EOS600D digital cameras were transferred wirelessly to a server and imported to the IMS through the drag and drop functionality to appear with the slides in the IMS reporting window. Understanding the HL7 message and educating all involved was crucial in the following steps.
Setting Up Integration Goals and Related Contracts
Prior to purchasing WSI system with its IMS, identifying the level of integration and setting it contractually is paramount in its achievement and limiting the cost. A barcode-only integration enabling data available within the barcode configuration (such as case number and stain) to be transferred to the IMS was considered but not applied due to its severe limitations.
Unidirectional LIS to IMS
During the purchasing process, Philips, our IMS provider, was under contractual obligation toachieve the highest level of integration possible within the limitations of the LIS acceptance of messages. This resulted in commitment in LIS to IMS integration, as stated in the contract arrangement, linked financially through delivery of equipment milestones and integration stages, which proved crucial in project delivery.
For this integration, all interface communication between the IMS and LIS was mapped and transferred via the Infor Cloverleaf, an HL7 integration engine, implemented on top of Cerner at the hospital to facilitate Cerner integration with different systems, as illustrated in Figure 1.
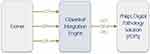 |
Figure 1 High level diagram of Cerner, Cloverleaf and IMS Communications. |
The patient data was sent to and updated in IMS via the HL7 messages provided by the LIS. Investigating and understanding available messages in the LIS (Cerner Pathnet) in order to enable transfer required collaborative work between the clinical and technical teams in order to decipher the messages and identify the trigger points and information required for transfer. An example of such a message (Figure 2) and its analysis are highlighted in Table 1.
 |
Table 1 Analysis of the HL7 Cloverleaf Message |
Integration Workflow
The workflow of the orders, related messages, tasks and reports required extensive involvement by all team members. The understanding of the lab and reporting process is paramount. In this, the involvement of an experienced lab technologist and anatomical pathology physician is essential. Time invested in this stage leads to reduced post-integration complications. The workflow, case movement and their trigger messages are highlighted in Figure 3 reporting a workflow with annotation of where an HL7 is created.
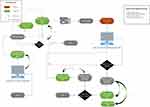 |
Figure 3 Full end-to-end integration workflow. |
Patient/Case Information Display in IMS
Integrated IMS information is displayed through utilization of HL7 messages. Multiple meetings were arranged in order to enable better understanding of the HL7 messages, defining clinical workflows, and trigger points between Cerner PathNet and IMS. A Cerner IT expert displayed the HL7 messages generated for each case and request scenario in order to identify the exact message components: patient identifiers (MRN, name, date of birth, age), slide status (requested, finalized, cancelled), slide type (number, stain, type), case status (requested, finished, cancelled, responsibility), report status (authorized primary report, addendum corrected report). The understanding of the individual HL7 message components allowed the Philips IMS team to integrate these messages in the IMS allowing appropriate display and actions. We managed to transfer and display the required targeted information: patient ID (created/managed by LIS), age, responsible reporting pathologists, stain type, tissue type, unique slide identifiers, periodization status (urgent, routine), slide order status (in preparation, finished) for each slide. The communication flow of all the HL7 messages with trigger events is illustrated in Figure 4.
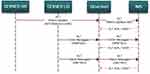 |
Figure 4 Communication flow of messages with trigger events. |
Case Movement Across Folders
Philips IMS in this aspect is locked and provides 5 folders: “In Preparation” for cases where slides are requested and not yet scanned or incompletely scanned; “Action Required” where slides require attention regarding barcode recognition; “For Review” where case is ready for the reporting pathologist; and “Finished” for reported cases, with a drop-down box listing the slides (Figure 5). The information regarding report status and slide status (ordered, authorized, cancelled) in the Cerner HL7 message were identified and used to transfer the cases across IMS folders. The slide-related messages were used to identify and list all slides requested, with case left “In Preparation” until all the slides are scanned, at which point the IMS will move the cases to the “For Review” folder in the responsible pathologist’s reporting list on the IMS. The cases were moved automatically from the “For Review” folder to the “Finished” folder using the observation result (ORU) triggered from the PathNet by finalizing the final and addendum reports, with frozen section and provisional report messages ignored because of the need to further view the slides and issue a report, as depicted in Figure 1. Cerner does not send any HL7 triggers when the responsible or reporting pathologist gets updated in PathNet. Due to this restriction, there was no way to report the pathologist updates to IMS in real time. In order to report the pathologist updates, the integration team wrote a CCL (Cerner Command Language) script to fetch reporting pathologist updates in CSV format (comma separated values), and push it to Cloverleaf. Once the CSV file is received at Cloverleaf integration engine, it transforms the CSV into HL7 message, and sends it to IMS. The CCL script is scheduled to run every 5 minutes through Cerner’s OPS (operational jobs) scheduler.
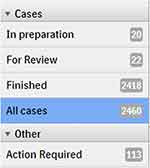 |
Figure 5 Folders in IMS. |
Integration Scripts and Test Plan
The integration workflow starts with the pathology tasks order entry (PTOE) in PathNet task creation of slide order. Ordering and saving a slide PTOE triggers the order message flows to the IMS “In Preparation” folder. The scanned slides are then coupled with the relevant slot in the IMS. Once all the case slides are scanned then the whole case is manually moved to the “For Review” folder once the quality control step is finalized. An additional slide request at this stage would move the case to the “In Preparation” folder. This function stops the pathologist unknowingly authorizing a case without viewing all the slides. There is an icon that shows when a slide has been reviewed in the IMS, also ensuring all scanned slides are examined. Authorizing the case in the LIS allows for a message to move the case to the finished folder. The integration was tested using validation scripts, allowing for testing of different case scenarios. Validation of integration scripts, testing every eventuality in these workflows, was performed, as illustrated in Table 2.
 |
Table 2 Validation of the integration scripts |
Recurrent Integration Errors and Deficiencies
Integration issues identified at quality assurance and reported to information technology for input peaked in the months after go live and were mainly related to two issues (Figure 6). First, processing task order entry (PTOE) tasks relating to newly introduced stains not being recognized as a trigger for sending the message to the IMS. Once identified, this was solved by introducing a change request procedure to include updating the stain list in the integration script, allowing these messages to be sent and recognized by the IMS. The second issue concerned the need to resend the message relating to the identification of a slide. Upon investigation, this was due to deletion and request of a new slide or test without saving each of these entries, leading to the two messages being received at the same time. A script was written to enable the integration to act on both these messages in the right order.
 |
Figure 6 Integration issues requiring integration team input. |
Discussion
Transforming into a fully digital anatomical pathology department had its own challenges. Integration between the LIS and the IMS represented the biggest challenge in the whole project. The limitation faced resulted from the lack of preexisting integration agreements between the vendors of the LIS and IMS. Pooling resources, knowledge and expertise from the IT integration and pathology teams enabled us to establish a satisfactory integration, enabling case management through the IMS in an IMS-driven workflow. The reporting pathologist is able view all cases with their related slides, extra work and pending requests in one platform (IMS), in addition to grossing, electron microscopy and other images being imported and available in the same platform. The integration making the report available as a PDF file allows for an availability of cases for review being instantly accessible.
There are multiple limitations. The reporting process achieved is still reliant on manually moving to the LIS in order to dictate the report. This shortfall allows for the possibility of an error by reporting the slides of one case under the LIS record of another patient, equivalent to picking up the wrong glass slides in a traditional laboratory. We mitigated this risk by contextually launching the IMS slides and LIS record of the case on two screens. We were not able to enable the reporting to be directly from the IMS. The same applies for requesting extra investigations. The main challenge for this bi-directional integration between Cerner Anatomical Pathology PathNet and the Philips Intellisite Pathology was the lack of contractual obligation from Cerner to engage in such integration.
Conclusions
Embarking on such integration, our experience would have been significantly improved and would have been smoother through the application of the following steps:
- Setting a clear contractual obligation for the integration requirement with the vendor from the outset.
- Understanding the workflow of physical slides and its relation with the virtual messages, early on.
- Setting clear integration goals, requiring extensive involvement of the medical staff from the outset. Here, we advise the involvement of as many staff as possible, in order to avoid new integration goals with the increased number of users.
- Identifying the possible changes in the LIS system and informing the integration team of such changes through change request reduces time processing such errors.
- Recognising the need for an increase in information technology support in the period following going live is an element to be considered in resource allocation during planning.
- Laying quality assurance measures prior to integration allows for a better process: developing validation scripts, testing scripts, document errors, change request.
Our experience highlighted a deficiency in the WSI pathology technology and its application as an integrated reporting platform. We recommend an industry-wide attempt to integrate the main WSI IMS platforms with the major LIS providers to cut the time and effort required for each institution going full digital. Such integration is essential for the technology to be a truly digital platform for reporting rather than another way to view the slides.
Data Sharing Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethical Approval and Consent to Participate
Not applicable. All data was treated within patient privacy and protection regulations.
Acknowledgments
Open Access funding provided by the Qatar National Library.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study project required no specific funding.
Disclosure
The authors declare that they have no competing interests in this work.
References
1. Snead DR, Tsang YW, Meskiri A, et al. Validation of digital pathology imaging for primary histopathological diagnosis. Histopathology. 2016;68(7):1063–1072. doi:10.1111/his.12879
2. Fraggetta F, Caputo A, Guglielmino R, et al. Guide for the rapid transition to a fully digital workflow: the “caltagirone example”. Diagnostics. 2021;11(10):1916. doi:10.3390/diagnostics11101916
3. Zarella MD, Bowman D, Aeffner F, et al. A practical guide to whole slide imaging: a white paper from the digital pathology association. Arch Pathol Lab Med. 2019;143(2):222–234. doi:10.5858/arpa.2018-0343-RA
4. Evans AJ, Bauer TW, Bui MM, et al. US Food and Drug Administration approval of whole slide imaging for primary diagnosis: a key milestone is reached and new questions are raised. Arch Pathol Lab Med. 2018;142(11):1383–1387. doi:10.5858/arpa.2017-0496-CP
5. Amin M, Sharma G, Parwani AV, et al. Integration of digital gross pathology images for enterprise-wide access. J Pathol Inform. 2012;3:10. doi:10.4103/2153-3539.93892
6. Fraggetta F, L’imperio V, Ameisen D, et al. Best practice recommendations for the implementation of a digital pathology workflow in the anatomic pathology laboratory by the European Society of digital and integrative pathology (ESDIP). Diagnostics. 2021;11(11):2167. doi:10.3390/diagnostics11112167
7. Cheng CL, Azhar R, Sng SH, et al. Enabling digital pathology in the diagnostic setting: navigating through the implementation journey in an academic medical centre. J Clin Pathol. 2016;69(9):784–792. doi:10.1136/jclinpath-2015-203600
8. García-Rojo M. International clinical guidelines for the adoption of digital pathology: a review of technical aspects. Pathobiology. 2016;83(2–3):99–109. doi:10.1159/000441192
9. Evans AJ, Salama ME, Henricks WH, Pantanowitz L. Implementation of whole slide imaging for clinical purposes: issues to consider from the perspective of early adopters. Arch Pathol Lab Med. 2017;141(7):944–959. doi:10.5858/arpa.2016-0074-OA
10. Williams BJ, Bottoms D, Clark D, Treanor D. Future-proofing pathology part 2: building a business case for digital pathology. J Clin Pathol. 2019;72(3):198–205. doi:10.1136/jclinpath-2017-204926
11. Volynskaya Z, Chow H, Evans A, Wolff A, Lagmay-Traya C, Asa SL. Integrated pathology informatics enables high-quality personalized and precision medicine: digital pathology and beyond. Arch Pathol Lab Med. 2018;142(3):369–382. doi:10.5858/arpa.2017-0139-OA
12. Aeffner F, Zarella MD, Buchbinder N, et al. Introduction to digital image analysis in whole-slide imaging: a white paper from the digital pathology association. J Pathol Inform. 2019;10:9. doi:10.4103/jpi.jpi_82_18
13. Robboy SJ, Gupta S, Crawford JM, et al. The pathologist workforce in the United States: II. An interactive modeling tool for analyzing future qualitative and quantitative staffing demands for services. Arch Pathol Lab Med. 2015;139(11):1413–1430. doi:10.5858/arpa.2014-0559-OA
14. Bick U, Lenzen H. PACS: the silent revolution. Eur Radiol. 1999;9(6):1152–1160. doi:10.1007/s003300050811
15. Mansoori B, Erhard KK, Sunshine JL. Picture archiving and communication system (PACS) implementation, integration & benefits in an integrated health system. Acad Radiol. 2012;19(2):229–235. doi:10.1016/j.acra.2011.11.009
16. van Diest PJ, Huisman A, van Ekris J, et al. Pathology image exchange: the Dutch digital pathology platform for exchange of whole-slide images for efficient teleconsultation, telerevision, and virtual expert panels. JCO Clin Cancer Inform. 2019;3:1–7. doi:10.1200/CCI.18.00146
17. Lechleitner G, Pfeiffer KP, Wilhelmy I, Ball M. Cerner Millennium™: the Innsbruck experience. Methods Inf Med. 2003;42(1):08–15. doi:10.1055/s-0038-1634204
 © 2023 The Author(s). This work is published by Dove Medical Press Limited, and licensed under a Creative Commons Attribution License.
The full terms of the License are available at http://creativecommons.org/licenses/by/4.0/.
The license permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
© 2023 The Author(s). This work is published by Dove Medical Press Limited, and licensed under a Creative Commons Attribution License.
The full terms of the License are available at http://creativecommons.org/licenses/by/4.0/.
The license permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

