Back to Journals » Journal of Inflammation Research » Volume 17
Discovery of mmu-lncRNA129814/hsa-lncRNA582795 as a Potential Biomarker and Intervention Target for Ischemia Reperfusion Injury-Induced AKI
Authors Yang L, Xu Y, Pan J, Li R, Lan C, Zhang D
Received 28 March 2024
Accepted for publication 22 June 2024
Published 2 July 2024 Volume 2024:17 Pages 4277—4296
DOI https://doi.org/10.2147/JIR.S465910
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Tara Strutt
Liu Yang,1 Yan Xu,1 Jian Pan,2– 4 Renjie Li,1 Chao Lan,1 Dongshan Zhang2– 4
1Department of Emergency Medicine, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan, People’s Republic of China; 2Department of Emergency Medicine, Second Xiangya Hospital, Central South University, Changsha, Hunan, People’s Republic of China; 3Emergency Medicine and Difficult Diseases Institute, Second Xiangya Hospital, Central South University, Changsha, Hunan, People’s Republic of China; 4Department of Nephrology, Second Xiangya Hospital, Central South University, Changsha, Hunan, People’s Republic of China
Correspondence: Chao Lan, Department of Emergency Medicine, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan, 450000, People’s Republic of China, Tel +86 13663855629, Email [email protected] Dongshan Zhang, Department of Emergency Medicine, Second Xiangya Hospital, Central South University, Changsha, Hunan, 410011, People’s Republic of China, Tel +86 138 7589 9625, Email [email protected]
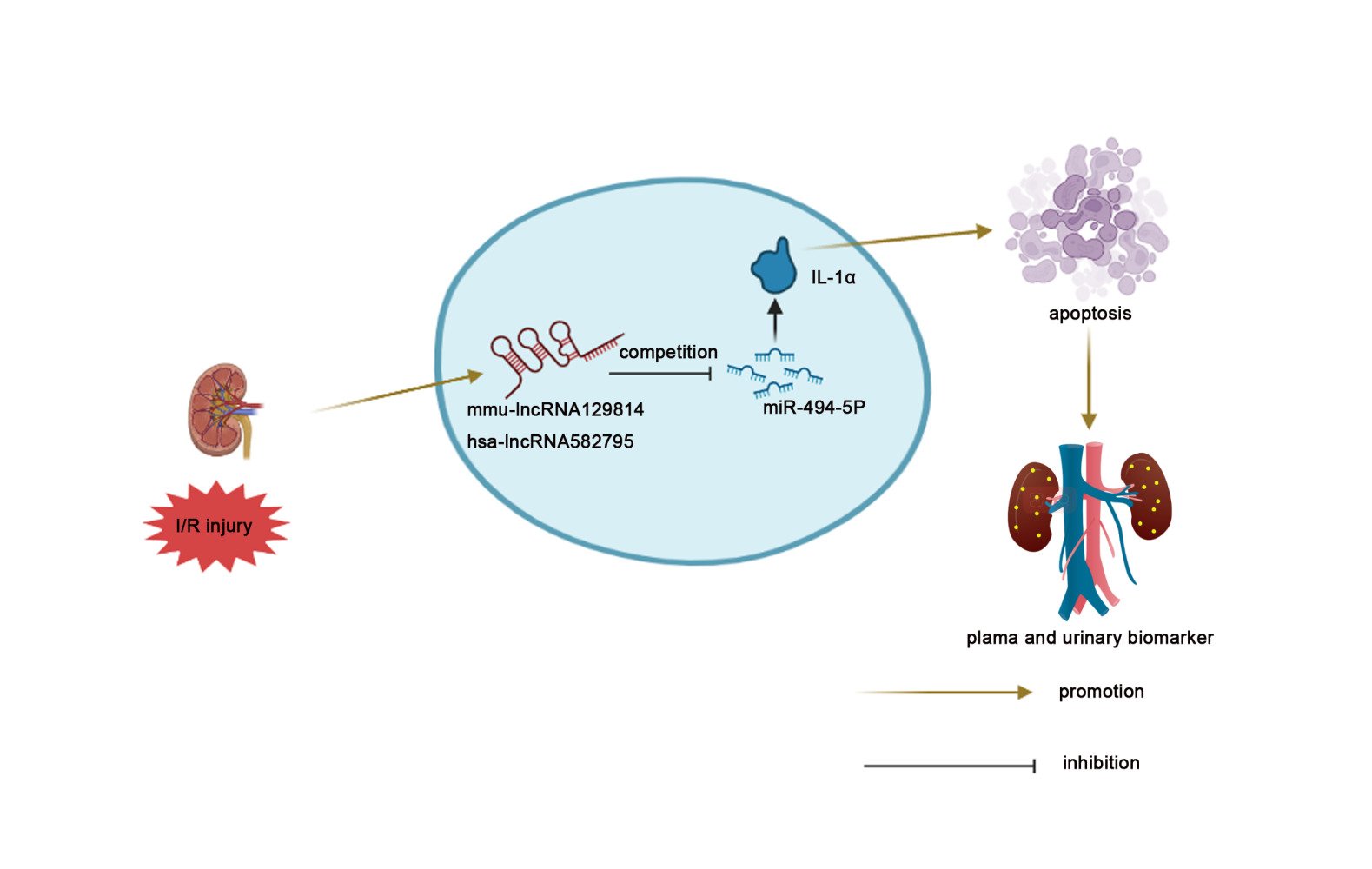
Background: Acute kidney injury (AKI) is associated with higher perioperative mortality and morbidity, as well as increased medical expenses. The molecular mechanisms underlying ischemia-reperfusion (I/R)-induced AKI remain unclear.
Methods and Results: We applied an RT-qPCR assay to measure the expression of mmu-lncRNA129814, hsa-lncRNA582795, and miRNA-494-5p, immunoblotting to detect IL-1α and cleaved caspase-3 expression, and TUNEL staining and flow cytometry (FCM) to evaluate apoptosis. The experiments were conducted using BUMPT and HK-2 cells, as well as C57BL/6J mice. Mechanistically, mmu-lncRNA129814 could sponge miRNA-494-5p and upregulate IL-1α expression to promote cell apoptosis. Furthermore, knockdown of mmu-lncRNA129814 ameliorated I/R-induced progression of AKI by targeting the miRNA-494-5p/IL-1α pathways. Interestingly, hsa-lncRNA582795, a homolog of mmu-lncRNA129814, also promoted I/R-stimulated HK-2 cell apoptosis and AKI progression by regulating the miRNA-494-5p/IL-1α axis. Finally, we found that patients with I/R-induced AKI exhibited significantly elevated plasma and urinary levels of hsa-lncRNA582795 compared to those who underwent ischemia-reperfusion without developing AKI. Spearman’s test demonstrated a significant correlation between serum creatinine and plasma hsa-lncRNA582795 in I/R patients. Plasma hsa-lncRNA582795 showed high sensitivity but low specificity (86.7%) compared to urinary hsa-lncRNA582795.
Conclusion: The mmu-lncRNA129814/hsa-lncRNA582795/miRNA-494-5p/IL-1α axis was found to modulate the progression of ischemic AKI, and hsa-lncRNA582795 could act as a diagnosis biomarker and potential therapy target of I/R-induced AKI.
Keywords: AKI, mmu-lncRNA129814, hsa-lncRNA582795, apoptosis, biomarker
Graphical Abstract:
Background
Acute kidney injury (AKI), a prevalent and serious complication, occurs in approximately 5–42% of 2 million cardiac surgeries annually.1–4 Ischemia-reperfusion (I/R)-induced AKI, which ranks as the first leading cause of AKI in the intensive care unit, is associated with high perioperative mortality and morbidity as well as increased medical expense.5,6 Although various factors such as inflammation, neurohormonal activation, oxidative stress, nephrotoxicity, and metabolic factors can contribute to the development of AKI, ischemia-reperfusion is still considered the primary cause.5,7 Over the past two decades, numerous signaling pathways have been identified as being related to the development of ischemia-reperfusion-induced AKI.8–10 However, the exact mechanism of this disease remains largely unknown. In addition, many biomarkers, such as alkaline phosphatase (AP), γ-glutamyl transpeptidase (GGT), KIM1, IL-18, NGAL, cystatin C (CST3C), liver fatty acid-binding protein (L-FABP), insulin-like growth factor-binding protein 7 (IGFBP7) and tissue inhibitor of metalloproteinases 2 (TIMP2), hold potential for the diagnosis of I/R-induced AKI.11–13 However, these biomarkers belong to protein markers for AKI.14,15 Therefore, it is of utmost urgency to discover a new nucleic acid biomarker for early diagnosis and treatment of I/R-induced AKI.
It has been recently found that long non-coding RNAs (lncRNAs), > 200 nucleotides, were involved in the progression of ischemic AKI.16 Some LncRNAs including SNHG14, eosinophil granule ontogeny transcript (EGOT), X–inactive specific transcript (XIST), GAS5,00520, MEG3, mmu-lncRNA 121686/hsa-lncRNA 520657, lncRNA 148400, and lncRNA147219 promoted the development of ischemic AKI.16–25 Other lncRNAs including lncRNA 00052, H19, TUG1, 00963, lncRNA 171502, lncRNA 122049, and lncRNA 136131 could alleviate cell apoptosis during ischemic AKI.4,26–31 Typically, lncRNAs sponged miRNAs to induce the target gene expression.32 However, whether these lncRNAs exhibit a potential diagnosis value for I/R-induced AKI remains largely unknown. We found that mmu-lncRNA129814 was localized at Chromosome 11: 108,528,241–108,751,079. However, its function, regulation mechanism, and potential diagnosis value in I/R-induced AKI are still unclear.
In the present study, mmu-lncRNA129814 sponged miRNA-494-5p to induce the expression of interleukin 1 alpha (IL-1α) and then mediate I/R-stimulated BUMPT cell death by modulating the miRNA-494-5p/IL-1α axis. Interestingly, hsa-lncRNA582795 exhibited the same function and regulatory mechanism with homologous mmu-lncRNA129814. Furthermore, the plasma and urinary levels of hsa-lncRNA 582795 could serve as a potential diagnosis biomarker for I/R-induced AKI.
Materials and Methods
Clinical Data and Specimens
We collected blood and urine samples from patients after obtaining ethical approval from the First Affiliated Hospital of Zhengzhou University (2022-KY-0270). The patients, who were treated with cardiopulmonary bypass during cardiac surgery, were enrolled between September 1, 2022, and April 30, 2023. Patients with previous renal disease, renal replacement therapy, malignancy, and taking nephrotoxic drugs were excluded. Patients who fell into the following categories were also excluded: under the age of 18, over the age of 80. All patients provided written informed consent. Healthy controls were collected from the physical examination department. Clinical data such as demographic characteristics, comorbidities, vital signs, type of surgery, operation time, cost, serum creatinine and blood urea nitrogen on admission were recorded. According to the combination status of acute kidney injury, these patients were assigned to CSA-AKI group (n = 30) and CSA-non-AKI group (n = 30). A healthy group served as the control (n = 15).
Antibodies and Regents
IL-1α (Cat#2170) was obtained from ABclonal company (Wuhan City, China). Anti-Cle-Casp3 (Cat#9661s) and Casp3 (Cat#9662s) were supplied by Cell Signaling Technology (MA, USA). Anti-tubB (Cat#T0023) was purchased from Affinity Biosciences (OH, USA). Lipofectamine 2000 (Cat#11668019) was supplied by Invitrogen (CA, USA). TRIzol reagent (Cat#AG21102), SYBR Green ProTaq HS premix (Cat#AG11701), and Evo M-MLV RT Mix Kit (Cat#AG11711) were obtained from Accurate Biology (Changsha, China). Trypsin-EDTA (Cat#C100C1) was purchased from NCM Biotech (Suzhou, China). HBSS (Cat#AWC0157a) was obtained from Changsha Abiowell Biotechnology (Changsha, China). Fluorescein isothiocyanate (FITC) and Annexin-V-FITC kit (Cat#556547) were obtained from BD Pharmingen (CA, USA). Dual-luciferase assay (Cat#E1910) was obtained from Promega (WI, USA). The mmu-lncRNA129814 siRNA, mmu-miRNA-494-5p mimic, mmu-miRNA-494-5p inhibitor and IL-1α siRNA were purchased from Ribo (Guangzhou, China). The mmu-lncRNA 129814 plasmid and hsa-lncRNA 582795 plasmid were purchased from Tsingke Biotechnology (Beijing, China).
Cell Culture, Ischemic Model, and Transfection
BUMPT and HK-2 cells were cultured in DMEM (Cat#C11995500BT, Gibco, Thermo, USA) and DMEM/F12 (Cat#1130500BT, Gibco, Thermo, USA) medium, respectively, containing 10% FBS (Cat#16000-044, Gibco, Thermo, USA) and 1% antibiotics (penicillin-streptomycin, Cat#15140122, Gibco, Thermo, USA) under a constant temperature of 37°C and 5% CO2. The BUMPT and HK-2 cells ischemia model was constructed with antimycin A (10 µM, Cat#MS0070, MK Biological technology, Shanghai, China) and calcium ionophore (1.5 µM, Cat#C346574, XI’an Aladdin Biological technology, China).16 The mmu-lncRNA129814 siRNA or plasmid, hsa-lncRNA 582795 plasmid, miRNA-494-5p mimic, miRNA-494-5p inhibitor, IL-1α siRNA or the negative control were transfected with Lip2000.
Ischemic AKI Mouse Model
Male C57BL/6J mice (8–10 weeks, weighted 20–25 g) were supplied by Hunan Slaike Jingda Laboratory Animal Company (Hunan, China), and gave adaptive feeding for 1 week. Before the ischemic surgery, the mmu-lncRNA 129814 siRNA (15 mg/kg) and hsa-lncRNA 582795 plasmid (25 ug) was administrated into each mouse through its tail vein once daily, and saline or vector was injected as control group. The renal pedicles were clamped bilaterally with arterial clips for 28 minutes and then subjected to reperfusion for 24 or 48 h.
Renal Function, Morphology, and Apoptosis
Blood specimens were withdrawn from the eyeballs of all mice at the end of the experiment flowed by the detection of renal function according to the protocols of BUN and sCr kits (Nanjing-Jiancheng-Bioengineering-Institute, Jiangsu, China). The left kidneys of all mice were removed and denatured with paraformaldehyde (4%) for 24 h. The sections of kidney were embedded in paraffin for Harris hematoxylin staining to assess the pathological changes of renal tissue. Histology was assessed by H&E staining based on the criteria for scoring renal tubular injury as previously described.16 Apoptosis was evaluated using TUNEL staining, following the methods outlined in previous studies. The quantitative measure of apoptosis was determined based on the percentage of TUNEL-positive cells in 10 to 20 microscopic fields per tissue section.16
qRT-PCR Assays
Trizol (AG, Changsha, China) was used to isolate total RNA from cells or tissues. Reverse transcription was carried out for cDNA synthesis in line with the instructions of Evo M-MLV RT Mix Kit (Cat#AG11711). PCR reaction system was set up based on the protocol of Ag SYBR Green Pro Taqhs premix (Cat#AG11701). The expression levels of mmu-lncRNA129814, hsa-lncRNA 582795, mmu-miRNA-494-5p, hsa-miRNA-494-5p, and IL-1α were examined by LightCycler® 480 II (Roche, Switzerland). The expression profiles of all lncRNAs in ischemic AKI were examined using a gene chip. Firstly, C57BL/6 mice were treated with I (28 min)/R (24h/48 h), and total RNA was extracted from the kidney cortex tissue for the lncRNA chip assay. Notably, the I/R group showed a significant upregulation of lncRNAs compared to the sham group. Among these, mmu-lncRNA 129814 (GenID: ENSMUST00000129814.8) was particularly upregulated at 48h. We used the UCSC Genome Browser to screen for human lncRNAs through gene conservation analysis. Ultimately, hsa-lncRNA 582795 (GenID: ENST00000582795.5) was identified through this conservative analysis. We found that both mmu-lncRNA 129814 and hsa-lncRNA 582795 contained the complementary sequences of several miRNAs via a target-prediction analysis software. And miR-494-5p is the only meaningful miRNA by qRT-PCR assay. Therefore, we focus on mmu-lncRNA129814, hsa-lncRNA582795 and miRNA-494-5p. The sequences of mmu-lncRNA 129814 and the hsa-lncRNA582795 were obtained from the Ensembl database (GenID:ENSMUST00000129814.8; Gen ID:ENST00000582795.5). The primer sequences were as follows: mmu-lncRNA129814: 5’- ACTCTCAGCGGAAACAGCAGTTG-3’ (F) and 5’-TTGACAGCAGTCTCATG GTCCTTTG-3’ (R); hsa-lncRNA582795.5:5’-AGGCGGCTCCAGGATGTTAGAC-3’ (F) and 5’-TTCTCTGCACATCGCACCTGATTAG-3’ (R); IL-1α:5’-TACAGTTCTGCCATTGACCATCT-3’ (F) and 5’-GTTGCTTGACGTTGCTGATACTG-3’ (R); β-actin: 5’-GGCTGTATTCCCCTCCATCG-3’ (F) and 5’-CCAGTTGGTAACAATGCCATGT-3’ (R); U6:5’-CTCGCTTCGGCAGCACA-3’ (F) and 5’-AACGCTTCACGAATTTGCGT-3’ (R). The primer sequences of miRNA-494-5p were supplied by Ribo bio (Guangzhou, China). The 2−∆∆CT method was applied for the above mentioned lncRNA or miRNA expression.
Immunoblot Analysis
The lysates from the renal cortex and whole cells were separated by SDS-PAGE and then transferred onto PVDF membranes. After blocking with 5% milk for 60 minutes, primary antibodies were applied and incubated overnight at 4°C. Following membrane washing, secondary antibodies were added and incubated for 60 minutes. Electrochemiluminescence (ECL) was used to detect the signals. Grayscale scanning was performed using a gel imaging analysis system.
Fluorescence in situ Hybridization (FISH)
The fluorescent probes of mmu-LncRNA129814, hsa-lncRNA582795, and miRNA-494-5p were designed and synthesized by GenePharma (Shanghai, China). FISH was carried out in line with the protocol of the in-situ hybridization kit (C10910, Ruibo).
Flow Cytometry (FCM) Analysis
The Annexin V/PI double staining was conducted to examine BUMPT and HK-2 cell death. Following digestion with EDTA-free trypsin, the cells were collected in a centrifuge tube, followed by washing with pre-cooled PBS twice and centrifugation at 4°C and 300 g for 5 min each time. The cell suspensions were added into the flow tube. FCM analysis was performed in accordance with the protocol of Annexin V kit (Cat#556547; BD Pharmingen, USA).
Statistical Analysis
Graphpad 9.5 and SPSS 25 were used for statistical analysis. The Student’s t-test was employed to compare the responses between two groups, while one-way ANOVA was utilized to compare the responses among multiple groups. The Kruskal-Walls test was employed for non-normally distributed data. The Chi-squared test was utilized for comparing categorical variables. The Spearman rank correlation coefficient was used to assess correlations among variables. Numerical data were expressed as mean ± standard error (SD). Sample means were compared with Mann–Whitney U-test. P < 0.05 was deemed statistically significant.
Results
Expression of Mmu-lncRNA129814 is Stimulated in BUMPT Cells and Mouse Kidneys After I/R
We examined whether I/R can affect the expression of mmu-lncRNA129814. Firstly, male C57BL/6J mice were treated with I (28 min)/R (24, 48 h). The serum concentrations of blood urea nitrogen (BUN) and creatinine (sCr) were elevated after reperfusion for 24 h and achieved a peak at 48 h (Figure 1A and B). These changes were consistent with the renal tubular injury degree assessed by both H&E staining and renal tubular damage scores (Figure 1C and D). The qRT-PCR assay revealed that mmu-lncRNA129814 expression was elevated after reperfusion for 24 h and achieved a peak at 48 h (Figure 1E), which is in line with the trend of the immunoblotting data of Cle-Casp3 (Figure 1F and G). BUMPT cells were incubated with calcium ionophore and antimycin A for 2 h and then subjected to reperfusion for 0, 2, and 4 h. The qRT-PCR assay showed that mmu-lncRNA129814 expression was remarkably elevated after reperfusion, achieved a peak at 2 h, and then decreased at 4 h (Figure 1H). This tend is consistent with the immunoblotting results of Cle-Casp3 (Figure 1I and J). Finally, FISH analysis indicated that mmu-lncRNA129814 was mainly localized in BUMPT cell cytoplasm (Figure 1K). Collectively, the data imply that mmu-lncRNA129814 expression can contribute to ischemic AKI progression.
Knockdown of Mmu-lncRNA129814 Attenuates I/R-Stimulated BUMPT Cell Death
We further investigated the effect of mmu-lncRNA129814 on BUMPT cell apoptosis during I/R. After transfection with mmu-lncRNA129814 siRNA, I(2-h)/R(2-h) treatment was carried out. The qRT-PCR assay demonstrated that mmu-lncRNA129814 siRNA inhibited its expression level under saline or I/R condition (Figure 2A). The FCM assessment demonstrated that I/R-stimulated BUMPT cell death was markedly inhibited by mmu-lncRNA129814 siRNA (Figure 2B and C). This effect was further verified by immunoblotting of Cle-Casp3 (Figure 2D and E). Collectively, these findings showed that mmu-lncRNA129814 was an apoptotic inducer during I/R.
Overexpression of Mmu-lncRNA129814 Enhances I/R-Stimulated BUMPT Cell Death
The siRNA targeting mmu-lncRNA129814 showed a reduction in cell death. Therefore, we proceeded to investigate whether overexpression of mmu-lncRNA129814 has the opposite effect. The qRT-PCR assay demonstrated that mmu-lncRNA129814 plasmid enhanced its expression level under saline or I/R condition (Figure 3A). In addition, mmu-lncRNA129814 could also enhance I/R-stimulated apoptosis in BUMPT cells, as examined by FCM and immunoblotting of Cle-Casp3 (Figure 3B–E). These data supported the conclusion that mmu-lncRNA129814 was an apoptosis inducer.
mmu-lncRNA 129814 Sponges miRNA-494-5p
Previous reports have demonstrated that lncRNAs can function as ceRNAs by binding to miRNAs. Through analysis with RegRNA v2.0, it has been found that mmu-lncRNA129814 contains complementary sequences to miRNA-494-5p (Figure 4A). It was found that the luciferase activity of mmu-lncRNA129814-WT, but not mmu-lncRNA129814-Mut, was noticeably suppressed by miRNA-494-5p mimic (Figure 4B). The FISH analysis showed that miRNA-494-5p and mmu-lncRNA129814 were interacted and co-localized in BUMPT cell cytoplasm and mouse kidneys under saline or sham and ischemic injury conditions (Figure 4C). Furthermore, qRT-PCR data revealed that knockdown of mmu-lncRNA 129814 could increase miRNA-494-5p expression under saline and ischemic injury conditions. In contrast, overexpression of mmu-lncRNA129814 inhibited its expression (Figure 4D and E). The data suggest that mmu-lncRNA129814 sponges miRNA-494-5p.
miRNA-494-5p Mimic Ameliorates BUMPT Cell Apoptosis Induced by I/R
Next, we assessed the effect of miRNA-494-5p on BUMPT cell death during ischemic injury. After transfection with miRNA-494-5p mimic, I(2-h)/R(2-h) treatment was carried out. The qRT-PCR assay showed that miRNA-494-5p mimic could increase miRNA-494-5p expression under saline or I/R condition (Figure 5A). The FCM evaluation indicated that I/R-stimulated BUMPT cell death was inhibited by the miRNA-494-5p mimic (Figure 5B and C). This effect was demonstrated by immunoblotting of Cle-Casp3 in BUMPT cells (Figure 5D and E). The data suggest that miRNA-494-5p can suppress I/R-stimulated BUMPT cell death.
IL-1α is a Target Gene of miRNA-494-5p
Despite its known anti-apoptotic function, the regulatory mechanism of miRNA-494-5p has remained elusive. However, through prediction analysis on the mirdb.org website, we identified IL-1α as a target gene of miRNA-494-5p. The sequence of IL-1α 3’UTR is complementary to miRNA-494-5p (Figure 6A). The luciferase reporter analysis showed that the luciferase activity of IL-1α3’ UTR-WT but not IL-1α3’ UTR-Mut was inhibited by miRNA-494-5p (Figure 6B). Furthermore, the miRNA-494-5p mimic was transfected into BUMPT cells, and then exposed to I(2-h)/R(2-h). The qRT-PCR assay demonstrated that miRNA-494-5p mimic markedly reduced IL-1α expression under saline or I/R condition (Figure 6C). This effect was further verified by immunoblotting of IL-1α (Figure 6D and E). These findings indicate that IL-1α can directly target miRNA-494-5p. To further investigate the effect of IL-1α on BUMPT cell death induced by I/R, IL-1α siRNA transfection and I(2-h)/R(2-h) exposure were conducted. The immunoblotting results demonstrated that IL-1α siRNA further suppressed I/R-stimulated upregulation of Cle-Casp3 (Figure 6F and G). These data imply that IL-1α is an apoptotic inducer during ischemic injury.
The Anti-Apoptotic Effect of mmu-lncRNA129814 siRNA on BUMPT Cells is Reversed by miRNA-494-5p Inhibitor During Ischemic Injury
To verify whether mmu-lncRNA129814 mediates apoptosis in BUMPT cells via sponging miRNA-494-5p, the miRNA-494-5p inhibitor and mmu-lncRNA19814 siRNA were co-transfected into BUMPT cells. The qRT-PCR assay revealed that mmu-lncRNA129814 siRNA inhibited mmu-lncRNA129814 expression, whereas transfection with miRNA-494-5p inhibitor suppressed the increased expression of miRNA-494-5p induced by mmu-lncRNA129814 siRNA during ischemic injury (Figure 7A and B). Both FCM and immunoblotting showed that knockdown of mmu-lncRNA 129814 markedly inhibited I/R-stimulated BUMPT cell death and reduced the expression of IL-1α and Cle-Casp3, and the miRNA-494-5p inhibitor could reverse these effects (Figure 7C–F). The data indicate that mmu-lncRNA 129814 can sponge miRNA-494-5p to induce BUMPT cell apoptosis.
Knockdown of Mmu-lncRNA129814 Ameliorates I/R Injury in Mice by Regulating the miRNA-494-5p/ IL-1α Axis
To clarify whether suppression of mmu-lncRNA129814 can prevent the progression of ischemic AKI, mmu-lncRNA129814 siRNA or saline was administrated into each mouse through its tail vein for 12 h prior to I(28 min)/R(48 h) exposure. The I/R-stimulated upregulation of BUN and sCr was markedly reduced by mmu-lncRNA129814 siRNA (Figure 8A and B). Consistently, H&E and TUNEL staining indicated that I/R-stimulated renal tubular injury and cell death were ameliorated by the mmu-lncRNA129814 siRNA (Figure 8C–F). The qRT-PCR assay demonstrated that mmu- lncRNA129814 siRNA inhibited its expression levels under sham or I/R condition (Figure 8G), however, it reversed the I/R-stimulated inhibition of miRNA-494-5p (Figure 8H). The immunoblotting indicated that I/R-stimulated upregulation of Cle-Casp3 and IL-1α was markedly ameliorated by mmu-lncRNA129814 siRNA (Figure 8I and J). Altogether, knockdown of mmu-lncRNA129814 protected against ischemic AKI by regulating I/R via the miRNA-494-5p/IL-1α axis.
I/R Induces Hsa-lncRNA 582795 Expression in HK-2 Cells
Hsa-lncRNA582795 (Ensemble: ENST00000582795.5) was identified from the Homo database (https://www.ncbi.nlm.nih.gov/). Interestingly, the common sequence between hsa-lncRNA 582795 and mmu-lncRNA129814 accounted for 63% of the mmu-lncRNA129814 sequence, while it comprised 86% of the hsa-lncRNA 582795 sequence (Figure S1A), suggesting a relatively high degree of homology between them. The qRT-PCR assay indicated that hsa-lncRNA 582795 expression was slightly elevated after reperfusion, achieved a peak at 2 h, and subsequently reduced at 4 h. This change was consistent with that of mmu-lncRNA129814 (Figure S1B). The immunoblotting result of Cle-Casp3 also aligned with the trend in hsa-lncRNA582795 expression during ischemic injury (Figure S1C and D). FISH analysis showed that hsa-lncRNA582795 was mainly localized in HK-2 cell cytoplasm (Figure S1E). In sum, these findings suggest that the expression of hsa-lncRNA582795 is increased in HK-2 cells following ischemic AKI.
Hsa-lncRNA 582795 Enhances I/R-Stimulated HK-2 Cell Death by Regulating the miRNA-494-5p/IL-1α Axis
To verify the effect of hsa-lncRNA 582795 on HK-2 cell death during ischemic AKI, transfection of hsa-lncRNA 582795 plasmid was conducted, followed by I(2-h)/R(2-h) exposure. The qRT-PCR assay demonstrated that hsa-lncRNA 582795 plasmid enhanced its expression levels under saline or I/R condition (Figure S2A). The FCM data indicated that overexpression of hsa-lncRNA 582795 could enhance I/R-stimulated HK-2 cell death (Figure S2B and C). The sequence of hsa-lncRNA 582795 is complementary to miRNA-494-5p; while that of miRNA-494-5p is complementary to IL-1α 3’UTR (Figure S2D). The qRT-PCR assay indicated that overexpression of hsa-lncRNA 582795 could suppress miRNA-494-5p expression under saline or I/R condition (Figure S2E). In contrast, the expression of IL-1α was increased by hsa-lncRNA 582795 overexpression under saline or I/R condition (Figure S2F). The immunoblotting results indicated that I/R-stimulated upregulation of Cle-Casp3 and IL-1α was further induced by hsa-lncRNA 582795 overexpression (Figure S2G and H). These findings demonstrate that hsa-lncRNA 582795 enhances I/R-stimulated HK-2 cell death via regulation of the miRNA-494-5p/IL-1α axis.
Overexpression of Hsa-lncRNA582795 Enhanced I/R Injury in Mice via Regulating the miRNA-494-5p/IL-1α Axis
To further demonstrate that hsa-lncRNA582795 has the same function as mm-lncRNA129814 in ischemic AKI, hsa-lncRNA582795 plasmid or vector was administrated into each mouse through its tail vein for 12 h before I(28 min)/R(48 h) exposure. I/R-stimulated upregulation of BUN and sCr was markedly increased by hsa-lncRNA582795 plasmid (Figure S3A and B). Consistently, H&E and TUNEL staining indicated that I/R-stimulated renal tubular injury and cell death were enhanced by hsa-lncRNA582795 plasmid (Figure S3C–F). The qRT-PCR assay demonstrated that hsa-lncRNA582795 plasmid enhanced its expression levels under sham or I/R condition (Figure S3G), Additionally, it aggravated the I/R-stimulated inhibition of miRNA-494-5p (Figure S3H). The immunoblotting results showed that the I/R-stimulated upregulation of Cle-Casp3 and IL-1α was markedly promoted by hsa-lncRNA582795 plasmid (Figure S3I and J). Taken together, overexpression of hsa-lncRNA582795 promotes ischemic AKI by regulating I/R via the miRNA-494-5p/ IL-1α axis.
hsa-lncRNA 582795 is a Diagnostic Marker of I/R-Induced AKI
It is well known that cardiac surgery can cause ischemic acute kidney injury (AKI). In this study, 60 post-cardiac surgery patients were enrolled, divided into the CSA-non-AKI group (n = 30) and the CSA-AKI group (n = 30), with a Healthy group (n = 15) serving as controls. Blood and urine samples were collected within 24 hours post-surgery. The baseline characteristics of all patients are presented in Table 1. Notably, the average ages of AKI and non-AKI patients were 55.8 ± 11.47 years and 46.83 ± 16.72 years, respectively. In AKI group, 10 (33.3%), 9 (30%) and 11 patients (66.7%) belonged to AKI stages 1, 2, and 3, respectively. The duration of cardiac surgery and the cost of hospitalization were higher in the AKI group compared to the non-AKI group (p < 0.05). To assess whether hsa-lncRNA 582795 was a diagnosis marker for cardiac surgery patients with ischemic AKI, total RNA was isolated from the blood and urine of healthy, non-AKI, and AKI groups (Figure S4A). The standard curve of absolute quantification PCR (AQ-PCR) was constructed using the hsa-lncRNA 582795 plasmid. Equation of regression is as follows: Y = −3.1136x + 35.434 (R = 0.9935) (Figure S4B). AQ-PCR assay indicated that the plasma and urinary levels of hsa-lncRNA582795 were slightly elevated in non-AKI patients compared with the healthy group, and those in AKI patients were noticeably increased compared to non-AKI patients (Figure S4C and D). Furthermore, the plasma and urinary concentrations of hsa-lncRNA 582795 were slightly increased in patients with Kidney Disease: Improving Global Outcomes (KDIGO) AKI stage II compared to those with KDIGO AKI stage I. In patients with KDIGO AKI stage III, these concentrations were markedly increased compared to those with KDIGO AKI stage II (Figure S4E and F). Additionally, the plasma and urinary levels of hsa-lncRNA 582795 were remarkably correlated with sCr (r = 0.824, p < 0.001; r = 0.734, p < 0.001, respectively) (Figure S4G and H). At the same time, we found that the elevated level of urinary hsa-lncRNA 58279 was consistent with the changing trend of plasma hsa-lncRNA 582795 (r = 0.816, p < 0.001) (Figure S4I). Our analysis confirmed that urinary hsa-lncRNA582795 had a strong correlation with urinary [TIMP2]*[IGFBP7](r = 0.789, p < 0.001;)(Figure S4J). To further evaluate the diagnostic value of hsa-lncRNA 582795, receiver operating characteristic (ROC) curves were established. As displayed in Figure S4K, the area under the ROC curve (AUC) of plasma hsa-lncRNA 582795 was 0.889 (95% CI, 0.808–0.970) with the sensitivity and specificity of 76.7% and 86.7%, respectively, at a cut-off value of 183.9×10−3 fg/L. The AUC of urinary hsa-lncRNA 582795 was 0.804 (95% CI, 0.693–0.916) with the lower sensitivity of 63.3% and higher specificity of 93.3% compared to its plasma level (Figure S4L). The data verify that hsa-lncRNA 582795 is an early marker for I/R-induced AKI during cardiac surgery.
 |
Table 1 The Baseline Clinical Characteristics and Laboratory Values of the Patients |
Discussion
This is the first study demonstrated that mmu-lncRNA129814/hsa-lncRNA 582795 promoted renal tubular cell apoptosis to drive the I/R-stimulated progression of AKI in cell and animal models. Mechanistically, mmu-lncRNA129814/hsa-lncRNA 582795 sponged miRNA-494-5p to increase the expression of IL-1α. Furthermore, the data suggested that plasma and urinary hsa-lncRNA 582795 had a potential diagnosis value for I/R-induced AKI patients.
Previous reports showed that lncRNAs could contribute to the development of ischemic AKI.16 Some lncRNAs protected against the progression of ischemic AKI,16–25 while others mediated the development of ischemic AKI.4,26–31 Here, we discovered that mmu-lncRNA 129814/hsa-lncRNA582795 not only mediated I/R-stimulated BUMPT and HK-2 cell death, but also aggravated the progression of ischemic AKI mice (Figures 2, 3, 8, S2 and S3). LncRNAs usually act as ceRNAs to promote the expression of target genes.33 It was observed that miRNA-494-5p was a target miRNA of mmu-lncRNA129814/hsa-lncRNA 582795 (Figures 4 and S2). Previous studies have also reported that miRNA-494-3p and miRNA-494-5p are two mature forms of miRNA-494. However, the effect of miRNA-494 on cell apoptosis remains controversial. Some studies have demonstrated that miRNA-494 mediated human nucleus pulposus and liver cell apoptosis.34,35 In contrast, other studies have found that miRNA-494 has the opposite effect on apoptosis in cardiomyocytes and vascular smooth muscle cells.36,37 The effect of miRNA-494-3p on apoptosis was similar with that of miRNA-494.38–40 However, the role of miRNA-494-5p in apoptosis is still unclarified. Herein, we reported for the first time that miRNA-494-5p had an anti-apoptotic function in renal tubular cell apoptosis during ischemic AKI (Figures 4, 5, 7, and 8). Our dual-luciferase reporter assays and qRT-PCR analysis showed that IL-1α was a target gene of miRNA-494-5p. Previous studies demonstrated that inhibition of IL-1α attenuated ischemic injury-induced myocardial injury and brain damage via suppression of apoptosis.41,42 Consistently, we also showed that the knockdown of IL-1α alleviated renal cell death induced by I/R (Figure 6). Interestingly, the findings showed that hsa-lncRNA 582795 was identical to mmu-lncRNA129814 and could modulate I/R-stimulated HK-2 cell death by regulating the miRNA-494-5p/IL-1α axis both in vitro and in vivo (Figures S2 and S3). In summary, the mmu-lncRNA129814/hsa-lncRNA 582795 miRNA-494-5p/IL-1α axis mediates the progression of I/R-induced AKI.
Early diagnosis of I/R-induced AKI can effectively improve the patient’s clinical outcome and mortality.43–46 Several studies reported that miRNA-21 had a diagnostic value for I/R-induced AKI. The area under the ROC curve ranged from 0.701 to 0.81 for plasma and from 0.68 to 0.9 for urine)\, respectively.47–49 However, these studies did not explore the sensitivity and specificity of miRNA-21 for diagnosing I/R-induced AKI. Recent studies suggest that lncRNAs belong to a novel class of diagnostic markers because they have tissue type-, cell type-, disease state- or developmental stage-specific patterns,50 which are easily detectable and highly stable in serum and urine.51 This is the first study indicated that hsa-lncRNA 582795 had a diagnosis value for I/R-induced AKI. The range of area under the ROC curve showed relatively high value of 0.889 (plasma) and moderate value of 0.804 (urine) compared to miRNA-21. Moreover, the data indicated that urinary hsa-lncRNA 582795 had a high specificity (93.3%) and low sensitivity (63.3%) compared to plasma hsa-lncRNA 582795 (86.7% and 76.7%, respectively). In addition, urinary hsa-lncRNA582795 was highly correlated with [TIMP2]*[IGFBP7] approved by FDA of Europe and the United States.52–54 This further confirms the ability of hsa-lncRNA582795 in diagnosing AKI (Figure S4). Collectively, these data show that hsa-lncRNA 582795 is a potential biomarker of I/R-induced AKI.
Conclusion
In summary, our study demonstrates that the mmu-lncRNA129814/hsa-lncRNA582795/miRNA-494-5p/IL-1α axis can contribute to the progression of I/R-induced AKI. The plasma and urinary hsa-lncRNA 582795 exhibit high specificity and sensitivity for I/R-induced AKI diagnosis, and may be a molecular target for treating I/R-induced AKI.
Abbreviations
BUMPT, Boston university mouse proximal tubule; AKI, Acute kidney injury; CABG, Coronary artery bypass graft; MAP, Mean arterial pressure; CSA-AKI, Cardiac surgery-associated AKI; I/R, Ischemia-reperfusion.
Data Sharing Statement
The datasets generated during and/or analysed during the current study are not publicly available due to [REASON(S) WHY DATA ARE NOT PUBLIC] but are available from the corresponding author on reasonable request].
Ethics Approval and Consent to Participate
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University (2022-KY-0270). The use of the BUMPT and HK-2 cell lines was approved by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University (2022-KY-0270). All animal experiments followed the guiding principles, which was approved by the Animal Care Ethics Committee of Second Xiangya Hospital, People Republic of China (NO.2018065).
Acknowledgments
We thank Changju Zhu and Zheng Dong for technical assistance. This paper has been uploaded to ResearchSquare as a preprint: https://www.researchsquare.com/article/rs-3832114/v1.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas, took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
The study was supported in part by a grant from National Natural Science Foundation of China (81870475). Changsha Science and Technology Bureau project (kq2001039). Key Project of Hunan provincial science and technology innovation (2020SK1014). Technology of Hunan Province project of International Cooperation and Exchanges (2020WK2009). The funding mentioned above funded the data collection for the study. The funds mentioned below funded the experimental reagents and the article writing. Fundamental Research China Hunan Provincial Science and Technology Department (2021SK4004). Henan Medical Science and Technology Research and Joint Construction Project (NO. LHGJ20200278, NO.LHGJ20220311), China Henan Provincial Science and Technology Department (212102310728).
Disclosure
The authors declare no competing interests in this work.
References
1. Demirjian S, Bashour CA, Shaw A, et al. Predictive accuracy of a perioperative laboratory test-based prediction model for moderate to severe acute kidney injury after cardiac surgery. JAMA. 2022;327(10):956–964. doi:10.1001/jama.2022.1751
2. Peng K, McIlroy DR, Bollen BA, et al. Society of cardiovascular anesthesiologists clinical practice update for management of acute kidney injury associated with cardiac surgery. Anesth Analg. 2022;135(4):744–756. doi:10.1213/ANE.0000000000006068
3. Tseng PY, Chen YT, Wang CH, et al. Prediction of the development of acute kidney injury following cardiac surgery by machine learning. Crit Care. 2020;24(1):478. doi:10.1186/s13054-020-03179-9
4. Li X, Zheng P, Ji T, Tang B, Wang Y, Bai S. LINC00052 ameliorates acute kidney injury by sponging miR-532-3p and activating the Wnt signaling pathway. Aging. 2020;13(1):340–350. doi:10.18632/aging.104152
5. Wang Y, Bellomo R. Cardiac surgery-associated acute kidney injury: risk factors, pathophysiology and treatment. Nat Rev Nephrol. 2017;13(11):697–711. doi:10.1038/nrneph.2017.119
6. Zarbock A, Küllmar M, Ostermann M, et al. Prevention of cardiac surgery-associated acute kidney injury by implementing the KDIGO guidelines in high-risk patients identified by biomarkers: the prevaki-multicenter randomized controlled trial. Anesth Analg. 2021;133(2):292–302. doi:10.1213/ANE.0000000000005458
7. Lameire N, Biesen WV, Vanholder R. Acute kidney injury. Lancet. 2008;372(9653):1863–1865. doi:10.1016/S0140-6736(08)61794-8
8. Ayad S, Neylan JF, Mayne TJ, Gouveia D, Swaminathan M. Hepatocyte growth factor mimetic ANG-3777 for cardiac surgery-associated acute kidney injury. Kidney Int Rep. 2020;5(12):2325–2332. doi:10.1016/j.ekir.2020.09.031
9. Stoppe C, Averdunk L, Goetzenich A, et al. The protective role of macrophage migration inhibitory factor in acute kidney injury after cardiac surgery. Sci Transl Med. 2018;10(441). doi:10.1126/scitranslmed.aan4886
10. Tiwari R, Kapitsinou PP. Role of endothelial prolyl-4-hydroxylase domain protein/hypoxia-inducible factor axis in acute kidney injury. Nephron. 2022;146(3):243–248. doi:10.1159/000518632
11. Endre ZH, Walker RJ, Pickering JW, et al. Early intervention with erythropoietin does not affect the outcome of acute kidney injury (the EARLYARF trial). Kidney Int. 2010;77(11):1020–1030. doi:10.1038/ki.2010.25
12. Kashani K, Al-Khafaji A, Ardiles T, et al. Discovery and validation of cell cycle arrest biomarkers in human acute kidney injury. Crit Care. 2013;17(1):R25. doi:10.1186/cc12503
13. Parikh CR, Thiessen-Philbrook H, Garg AX, et al. Performance of kidney injury molecule-1 and liver fatty acid-binding protein and combined biomarkers of AKI after cardiac surgery. Clin J Am Soc Nephrol. 2013;8(7):1079–1088. doi:10.2215/CJN.10971012
14. Koyner JL, Parikh CR. Clinical utility of biomarkers of AKI in cardiac surgery and critical illness. Clin J Am Soc Nephrol. 2013;8(6):1034–1042. doi:10.2215/CJN.05150512
15. Vandenberghe W, Van Laethem L, Herck I, et al. Prediction of cardiac surgery associated - acute kidney injury (CSA-AKI) by healthcare professionals and urine cell cycle arrest AKI biomarkers [TIMP-2]*[IGFBP7]: a single center prospective study (the PREDICTAKI trial). J Crit Care. 2022;67:108–117. doi:10.1016/j.jcrc.2021.10.015
16. Pan J, Xie Y, Li H, et al. mmu-lncRNA 121686/hsa-lncRNA 520657 induced by METTL3 drive the progression of AKI by targeting miR-328-5p/HtrA3 signaling axis. Mol Ther. 2022;30(12):3694–3713. doi:10.1016/j.ymthe.2022.07.014
17. Xue Q, Yang L, Wang H, Han S, Ramoji K. Silence of long noncoding RNA SNHG14 alleviates ischemia/reperfusion-induced acute kidney injury by regulating miR-124-3p/MMP2 axis. Biomed Res Int. 2021;2021:8884438. doi:10.1155/2021/8884438
18. Wang IK, Palanisamy K, Sun KT, et al. The functional interplay of lncRNA EGOT and HuR regulates hypoxia-induced autophagy in renal tubular cells. J Cell Biochem. 2020;121(11):4522–4534. doi:10.1002/jcb.29669
19. Chen F, Hu Y, Xie Y, et al. Total glucosides of paeony alleviate cell apoptosis and inflammation by targeting the long noncoding RNA XIST/MicroRNA-124-3p/ITGB1 axis in renal ischemia/reperfusion injury. Mediators Inflamm. 2020;2020:8869511. doi:10.1155/2020/8869511
20. Tang B, Li W, Ji T, et al. Downregulation of XIST ameliorates acute kidney injury by sponging miR-142-5p and targeting PDCD4. J Cell Physiol. 2020;235(11):8852–8863. doi:10.1002/jcp.29729
21. Geng X, Song N, Zhao S, et al. LncRNA GAS5 promotes apoptosis as a competing endogenous RNA for miR-21 via thrombospondin 1 in ischemic AKI. Cell Death Discov. 2020;6(1):19. doi:10.1038/s41420-020-0253-8
22. Tian X, Ji Y, Liang Y, Zhang J, Guan L, Wang C. LINC00520 targeting miR-27b-3p regulates OSMR expression level to promote acute kidney injury development through the PI3K/AKT signaling pathway. J Cell Physiol. 2019;234(8):14221–14233. doi:10.1002/jcp.28118
23. Liu D, Liu Y, Zheng X, Liu N. c-MYC-induced long noncoding RNA MEG3 aggravates kidney ischemia-reperfusion injury through activating mitophagy by upregulation of RTKN to trigger the Wnt/β-catenin pathway. Cell Death Dis. 2021;12(2):191. doi:10.1038/s41419-021-03466-5
24. Li X, Wu Z, Yang J, Zhang D. LncRNA 148400 promotes the apoptosis of renal tubular epithelial cells in ischemic AKI by targeting the miR-10b-3p/GRK4 axis. Cells. 2022;11(24):3986. doi:10.3390/cells11243986
25. Liu J, Li X, Yang J, Zhang D. LncRNA ENSMUST_147219 mediates the progression of ischemic acute kidney injury by targeting the miR-221-5p/IRF6 axis. Apoptosis. 2022;27(7–8):531–544. doi:10.1007/s10495-022-01730-3
26. Haddad G, Kölling M, Wegmann UA, et al. Renal AAV2-mediated overexpression of long non-coding RNA H19 attenuates ischemic acute kidney injury through sponging of microRNA-30a-5p. J Am Soc Nephrol. 2021;32(2):323–341. doi:10.1681/ASN.2020060775
27. Chen L, Xu JY, Tan HB. LncRNA TUG1 regulates the development of ischemia-reperfusion mediated acute kidney injury through miR-494-3p/E-cadherin axis. J Inflamm. 2021;18(1):12. doi:10.1186/s12950-021-00278-4
28. Xie LB, Chen B, Liao X, et al. LINC00963 targeting miR-128-3p promotes acute kidney injury process by activating JAK2/STAT1 pathway. J Cell Mol Med. 2020;24(10):5555–5564. doi:10.1111/jcmm.15211
29. Xu J, Wang B, Zhang D. LncRNA ENSMUST00000171502 induced by HIF-1α ameliorates ischemic acute kidney injury via targeting the miR-130b-3p/Mybl-1 axis. Cells. 2022;11(23):3747. doi:10.3390/cells11233747
30. Nagulakonda S, Pandey NN, Arvind B, Kumar S. Levoatriocardinal vein in a patient with transposition of great arteries and pulmonary stenosis. J Card Surg. 2022;37(9):2837–2838. doi:10.1111/jocs.16682
31. Wu Z, Pan J, Yang J, Zhang D. LncRNA136131 suppresses apoptosis of renal tubular epithelial cells in acute kidney injury by targeting the miR-378a-3p/Rab10 axis. Aging. 2022;14(8):3666–3686. doi:10.18632/aging.204036
32. Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22(2):96–118. doi:10.1038/s41580-020-00315-9
33. Xu J, Xu J, Liu X, Jiang J. The role of lncRNA-mediated ceRNA regulatory networks in pancreatic cancer. Cell Death Discov. 2022;8(1):287. doi:10.1038/s41420-022-01061-x
34. Wang T, Li P, Ma X, et al. MicroRNA-494 inhibition protects nucleus pulposus cells from TNF-α-induced apoptosis by targeting JunD. Biochimie. 2015;115:1–7. doi:10.1016/j.biochi.2015.04.011
35. Li H, Zhang L, Cai N, Zhang B, Sun S. MicroRNA-494-3p prevents liver fibrosis and attenuates hepatic stellate cell activation by inhibiting proliferation and inducing apoptosis through targeting TRAF3. Ann Hepatol. 2021;23:100305. doi:10.1016/j.aohep.2021.100305
36. Ning S, Li Z, Ji Z, et al. MicroRNA‑494 suppresses hypoxia/reoxygenation‑induced cardiomyocyte apoptosis and autophagy via the PI3K/AKT/mTOR signaling pathway by targeting SIRT1. Mol Med Rep. 2020;22(6):5231–5242. doi:10.3892/mmr.2020.11636
37. Cui R, Ye S, Zhong J, et al. MicroRNA‑494 inhibits apoptosis of murine vascular smooth muscle cells in vitro. Mol Med Rep. 2019;19(5):4457–4467. doi:10.3892/mmr.2019.10085
38. Wu C, Yang J, Li R, Lin X, Wu J, Wu J. LncRNA WT1-AS/miR-494-3p regulates cell proliferation, apoptosis, migration and invasion via PTEN/PI3K/AKT signaling pathway in non-small cell lung cancer. Onco Targets Ther. 2021;14:891–904. doi:10.2147/OTT.S278233
39. Wang H, Wang S, Huang S. MiR-494-3p alleviates acute lung injury through regulating NLRP3 activation by targeting CMPK2. Biochem Cell Biol. 2021;99(3):286–295. doi:10.1139/bcb-2020-0243
40. Hua J, Wang X, Ma L, et al. CircVAPA promotes small cell lung cancer progression by modulating the miR-377-3p and miR-494-3p/IGF1R/AKT axis. Mol Cancer. 2022;21(1):123. doi:10.1186/s12943-022-01595-9
41. Mauro AG, Mezzaroma E, Torrado J, et al. Reduction of myocardial ischemia-reperfusion injury by inhibiting interleukin-1 alpha. J Cardiovasc Pharmacol. 2017;69(3):156–160. doi:10.1097/FJC.0000000000000452
42. Liberale L, Bonetti NR, Puspitasari YM, et al. Postischemic administration of IL-1α neutralizing antibody reduces brain damage and neurological deficit in experimental stroke. Circulation. 2020;142(2):187–189. doi:10.1161/CIRCULATIONAHA.120.046301
43. Wen Y, Parikh CR. The aftermath of AKI: recurrent AKI, acute kidney disease, and CKD progression. J Am Soc Nephrol. 2021;32(1):2–4. doi:10.1681/ASN.2020091317
44. Koyner JL. Subclinical acute kidney injury is acute kidney injury and should not be ignored. Am J Respir Crit Care Med. 2020;202(6):786–787. doi:10.1164/rccm.202006-2239ED
45. Kellum JA, Romagnani P, Ashuntantang G, Ronco C, Zarbock A, Anders HJ. Acute kidney injury. Nat Rev Dis Primers. 2021;7(1):52. doi:10.1038/s41572-021-00284-z
46. Huang CT, Liu KD. Exciting developments in the field of acute kidney injury. Nat Rev Nephrol. 2020;16(2):69–70. doi:10.1038/s41581-019-0241-5
47. Du J, Cao X, Zou L, et al. MicroRNA-21 and risk of severe acute kidney injury and poor outcomes after adult cardiac surgery. PLoS One. 2013;8(5):e63390.
48. Gaede L, Liebetrau C, Blumenstein J, et al. Plasma microRNA-21 for the early prediction of acute kidney injury in patients undergoing major cardiac surgery. Nephrol Dial Transplant. 2016;31(5):760–766. doi:10.1093/ndt/gfw007
49. Arvin P, Samimagham HR, Montazerghaem H, Khayatian M, Mahboobi H, Ghadiri Soufi F. Early detection of cardiac surgery‑associated acute kidney injury by microRNA-21. Bratisl Lek Listy. 2017;118(10):626–631. doi:10.4149/BLL_2017_120
50. Liu S, Cao Q, An G, Yan B, Lei L. Identification of the 3-lncRNA signature as a prognostic biomarker for colorectal cancer. Int J Mol Sci. 2020;21(24):9359. doi:10.3390/ijms21249359
51. Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol. 2021;220(2). doi:10.1083/jcb.202009045
52. Pilarczyk K, Edayadiyil-Dudasova M, Wendt D, et al. Urinary [TIMP-2]*[IGFBP7] for early prediction of acute kidney injury after coronary artery bypass surgery. Ann Intensive Care. 2015;5(1):50. doi:10.1186/s13613-015-0076-6
53. Wang Y, Zou Z, Jin J, et al. Urinary TIMP-2 and IGFBP7 for the prediction of acute kidney injury following cardiac surgery. BMC Nephrol. 2017;18(1):177. doi:10.1186/s12882-017-0592-8
54. Titeca-Beauport D, Daubin D, Chelly J, et al. The urine biomarkers TIMP2 and IGFBP7 can identify patients who will experience severe acute kidney injury following a cardiac arrest: a prospective multicentre study. Resuscitation. 2019;141:104–110. doi:10.1016/j.resuscitation.2019.06.008
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.








