Back to Journals » Drug Design, Development and Therapy » Volume 18
Exploration of Berberine Against Ulcerative Colitis via TLR4/NF-κB/HIF-1α Pathway by Bioinformatics and Experimental Validation
Authors Li J , Dan W , Zhang C, Liu N, Wang Y , Liu J, Zhang S
Received 7 March 2024
Accepted for publication 25 June 2024
Published 9 July 2024 Volume 2024:18 Pages 2847—2868
DOI https://doi.org/10.2147/DDDT.S436359
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Frank Boeckler
Jilei Li,1,* Wenchao Dan,2,* Chenchen Zhang,3 Nian Liu,3 Yichong Wang,1 Jixiang Liu,3 Shengsheng Zhang1
1Digestive Disease Center, Beijing Hospital of Traditional Chinese Medicine, Capital Medical University, Beijing, 100010, People’s Republic of China; 2Department of Dermatology, Beijing Hospital of Traditional Chinese Medicine, Capital Medical University, Beijing, 100010, People’s Republic of China; 3Beijing University of Chinese Medicine, Beijing, 100029, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Shengsheng Zhang, Digestive Disease Center, Beijing Hospital of Traditional Chinese Medicine, Capital Medical University, Beijing, 100010, Tel +8613801088329, Email [email protected]
Purpose: This study aimed to delineate the molecular processes underlying the therapeutic effects of berberine on UC by employing network pharmacology tactics, molecular docking, and dynamic simulations supported by empirical validations both in vivo and in vitro.
Patients and Methods: We systematically screened potential targets and relevant pathways affected by berberine for UC treatment from comprehensive databases, including GeneCards, DisGeNET, and GEO. Molecular docking and simulation protocols were used to assess the interaction stability between berberine and its principal targets. The predictions were validated using both a DSS-induced UC mouse model and a lipopolysaccharide (LPS)-stimulated NCM460 cellular inflammation model.
Results: Network pharmacology analysis revealed the regulatory effect of the TLR4/NF-κB/HIF-1α pathway in the ameliorative action of berberine in UC. Docking and simulation studies predicted the high-affinity interactions of berberine with pivotal targets: TLR4, NF-κB, HIF-1α, and the HIF inhibitor KC7F2. Moreover, in vivo analyses demonstrated that berberine attenuates clinical severity, as reflected by decreased disease activity index (DAI) scores, reduced weight loss, and mitigated intestinal inflammation in DSS-challenged mice. These outcomes include suppression of the proinflammatory cytokines IL-6 and TNF-α and downregulation of TLR4/NF-κB/HIF-1α mRNA and protein levels. Correspondingly, in vitro findings indicate that berberine decreases cellular inflammatory injury and suppresses TLR4/NF-κB/HIF-1α signaling, with notable effectiveness similar to that of the HIF-1α inhibitor KC7F2.
Conclusion: Through network pharmacology analysis and experimental substantiation, this study confirmed that berberine enhances UC treatment outcomes by inhibiting the TLR4/NF-κB/HIF-1α axis, thereby mitigating inflammatory reactions and improving colonic pathology.
Keywords: berberine, ulcerative colitis, TLR4/NF-κB/HIF-1α, network pharmacology, molecular docking, experimental validation, inhibitor
Graphical Abstract:
Introduction
Ulcerative colitis (UC) is an inflammatory bowel disease characterized by symptoms such as abdominal pain, diarrhea, and the presence of blood and mucus in the stool. UC predominantly affects the mucosal and submucosal layers from the rectum to the colon, and its incidence is notably higher in regions such as South America and Europe.1 Concurrently, there have been notable increases in China, corresponding with economic growth and advancements in diagnostic technologies.2 UC is renowned for its chronic and relapsing nature, posing a substantial threat to patients’ well-being and contributing to the socioeconomic burden. The etiology of UC is complex and multifactorial, with environmental influences, genetic predispositions, microbial compositions, and immune responses all being implicated.3
UC treatment usually involves 5-ASA, corticosteroids, immunosuppressive drugs, and biological therapies. These interventions can ameliorate symptoms; however, challenges such as recurrence, suboptimal response rates, and significant costs persist.4,5 Recent studies have advocated the use of traditional Chinese medicine and its bioactive components, with evidence suggesting that these can reduce recurrence, minimize side effects, and be economically viable.6–9 Berberine, in particular, has been highlighted for its purported efficacy.10–12 Pharmacological research indicates that berberine may treat UC by restoring tight junction proteins in the intestine, modifying the gut microbiota, regulating immune function, repairing the intestinal barrier, and decreasing proinflammatory cytokines.13–16 There is evidence suggesting that TLR4 may invoke NF-κB activation in UC, leading to further intestinal inflammation and vascular injury while also stimulating HIF-1α expression, thus exacerbating UC pathology.17,18 Conversely, berberine has been suggested to aggravate UC by interfering with immune homeostasis and altering the tissue microenvironment, potentially through the action of hypoxia-inducible factor (HIF).19 However, determining whether berberine can beneficially modulate TLR4, NF-κB, and HIF-1α in UC requires additional investigation.
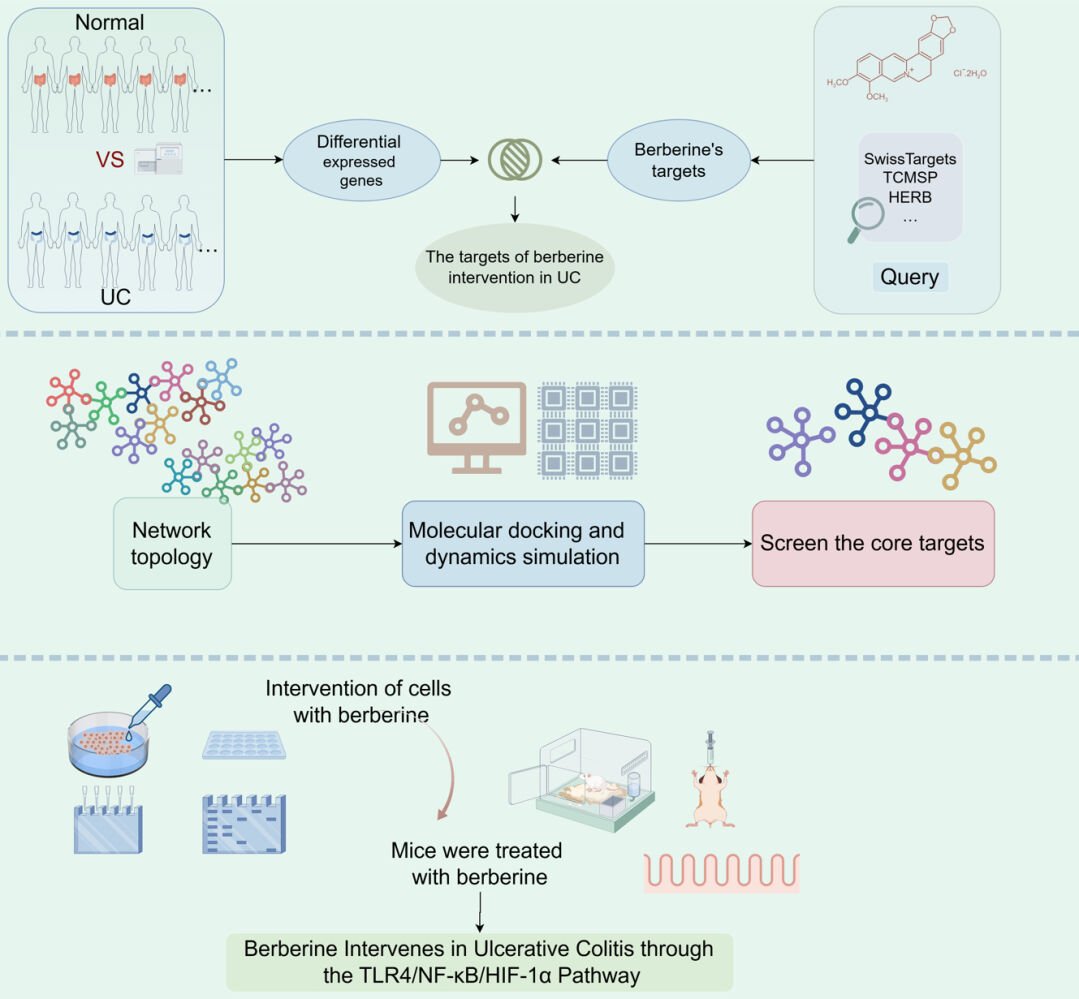
By applying network pharmacology—a support tool in drug discovery—this research offers an innovative methodology for predicting the complex interplay among drug components, biological targets, and disease states. Utilizing the Molecular Docking Method (MDM) and Molecular Dynamics (MD), this study employs computational simulations to active sites of drug-target interactions and ascertain the most favorable conformational alignments of ligands and receptors.20 Our approach begins with bioinformatic prediction and preliminary validation of potential targets and pathways associated with the action of berberine in UC treatment. Subsequent validation through in vivo mouse models and in vitro cellular assays will further explore the molecular pathways through which berberine may exert its therapeutic effects. A comprehensive workflow of the study protocol is depicted in Figure 1.
 |
Figure 1 The workflow of the analysis procedures. |
Materials and Methods
Network Pharmacology Analysis
Collection of Berberine Targets and Ulcerative Colitis Targets
We systematically retrieved potential targets of BBR by comprehensively searching multiple databases. Targets with a probability greater than or equal to 0.75 were extracted from the Super-PRED database, whereas the PharmMapper database contributed targets with a NormalFit score above 0.7.21–23 The SwissTargetPrediction platform was queried for targets with a probability threshold of 0.1 or higher. Furthermore, database searches were extended to HERB, ETCM, STCIH, TCMSP, and DrugBank to encompass a broad spectrum of BBR targets.24–28
Acquisition of Information on Related Pathways
Differential gene expression analysis between normal and UC tissue specimens conducted on the GSE107499 dataset from the GEO database yielded potential UC-associated targets. Additional disease-associated targets were accessed from the GeneCards and DisGeNET databases. To ascertain the current relevance of the findings, a manual literature review covering the most recent five years was performed, targeting publications delineating the association between BBR and UC for additional target identification. Subsequent standardization of these intersecting targets was carried out using the UniProt database.29
Protein-Protein Interaction (PPI) and Core Network Construction
The screened intersecting targets were imported into STRING 15.0 to generate a protein-protein interaction network model, in which ‘Homo sapiens’ was the biological species and the interaction confidence score was set to “highest confidence” (>0.9). Default parameters were otherwise maintained to construct the PPI network.30
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analysis
Key targets were predicted through topological network analysis using CytoScape 3.10. The Metascape platform was used for enrichment analysis of BBR targets implicated in UC, utilizing a significance threshold of P<0.01. The principal biological processes and metabolic pathways involved were systematically analyzed and visualized utilizing version 4.3.1 of the R software.
Molecular Docking Method
Building upon the comprehensive enrichment analysis and key biomarkers identified in the HIF pathway, we selected TLR4, TNF-α, HIF-1α, IL-1β, IL-6, IL-10, and NF-κB as receptor targets for molecular docking studies. BBR, along with the HIF-1α inhibitor KC7F2, was our ligand of interest. The three-dimensional crystal structures of these target proteins were retrieved in PDB from the RCSB database.31 Using Open Babel 3.1.1, we converted the 3D chemical structures of BBR and KC7F2 into the PDB format. The modified structures were saved as PDBQT files using AutoDock Tools 1.5.6.32 Guided by MOE2020, the molecular docking procedure was executed. The active components that displayed optimal binding affinity were identified by their docking scores. Specifically, a docking affinity score less than −4.25 kcal/mol was indicative of noteworthy binding activity, with scores less than −5 kcal/mol indicating strong binding potential.33–35
Molecular Dynamics
We performed molecular dynamics simulations of the ligand-protein complexes using Desmond 2020. The OPLS3e force field and TIP3 water model were used for solvation in the MD setup. Equilibration occurred in an NPT ensemble at 300 K and 1.0 bar using Berendsen coupling for temperature and pressure stabilization. After system preparation, a production run spanning 200 nanoseconds was initiated with a timestep of 1.2 femtoseconds, while the trajectories were captured at 200-picosecond intervals, aggregating to a total of 10,000 frames. The root mean square fluctuation (RMSF) calculations for each residue provided insight into the substantive conformational shifts, differentiating between the residue’s baseline configuration and its dynamic state.
Experimental Animals
Mouse Model of DSS-Induced Colitis and BBR Treatment
For this study, thirty-two specific pathogen-free male C57BL/6 mice aged between 6 and 8 weeks with an average weight of approximately 20 grams were employed. The animals were procured from SPF (Beijing) Biotechnology Co., Ltd. and housed under controlled conditions at the Beijing Institute of Traditional Chinese Medicine. Ethical approval for the research was granted by the Beijing Institute of Traditional Chinese Medicine Research Ethics Committee (BJTCM-M-2023-11-07). We also follow the “Laboratory Animal—Guideline for Ethical Review of Animal Welfare (GB/T 35892–2018)”. The mice were allocated randomly into four equal groups—namely, control, model, berberine-treated, and mesalazine-treated—utilizing a random number table method. The established protocol for inducing ulcerative colitis involved administering 3% DSS in the drinking water ad libitum for seven days.36 Pharmacological intervention, initiated on day four of post-colitis induction, was maintained for an additional seven days. Berberine (supplied by Jiang Lai Biotechnology, 02069–5 g) and mesalazine (procured from Kuihua Pharmaceutical, H19980148) were administered to the corresponding cohorts daily via gavage at a dosage of 100 mg/(kg·d). The placebo groups received an equivalent volume of saline (0.2 mL) following the same schedule.
DAI and Anal Condition
Observe and record the weight loss rate, blood in the stool, and stool consistency condition of mice daily, and score each item. The average of the three scores is the DAI score. The DAI score reflects the extent of colon tissue damage; the higher the score, the more severe the colon tissue damage. The specific criteria of the DAI score include included weight loss (scored as 0 for stable weight; 1 for 1–5% loss; 2 for 5–10% loss; 3 for 10–15% loss; 4 for more than 15% reduction), stool consistency (0 for normal; 2 for loose stools; 4 for diarrhea), and blood in the stool (0 for negative; 2 for occult blood; 4 for visible bleeding). After treatment, the mice’s anal region Condition was inspected and imaged.
Colon Length, Colon Mucosa Damage Index (CMDI), and Splenic Index
In the process of assessing colonic tissue and associated pathology, mice were dissected along the abdominal median line from the anus to the caecum to excise the colon for length measurements. Subsequently, the colonic tissue was opened, and the degree of mucosal damage was visually appraised and quantified according to the CMDI scoring system. The spleen was also removed and weighed for analysis. The CMDI score is delineated by the following scale: 0 indicates no mucosal damage; 1 is assigned for a smooth mucosal surface that exhibits mild congestion and edema in the absence of erosion or ulceration; a score of 2 corresponds to the mucosa with congestion, edema, coarseness, granularity, erosion, or adhesions; a score of 3 is attributed to the mucosal surface manifesting necrosis, ulceration, significant congestion, and edema with ulcer dimensions under 1.0 cm, coupled with necrosis, inflammation, or proliferation on the intestinal wall; a score of 4 is reserved for cases where the largest ulcer exceeds 1.0 cm or where full-thickness wall necrosis and inflammation are exceedingly severe.37 The splenic index reflects the inflammatory status of mice; the higher the splenic index, the more severe the inflammation. Record the body weight of the mice before sampling, remove and weigh the spleen at the time of sampling, and then proceed with the calculation using the formula below. Splenic index = (spleen weight (mg)/body weight (g)) × 10.
Haematoxylin and Eosin (H&E) Staining
Tissues from the colon, spleen, and lungs were fixed, dehydrated with alcohol, and embedded. The sections were deparaffinized, stained with hematoxylin and eosin, washed, and then mounted. Pathological alterations were evaluated using an inverted microscope to determine morphological changes.
Immunohistochemistry (IHC)
Paraffin-embedded colon tissue sections were deparaffinized and rehydrated before antigen retrieval in sodium citrate buffer solution (pH 6.0). The sections were subsequently incubated with primary antibodies against IL-6 and TNF-α overnight, visualized using DAB reagent for 1 minute, and counterstained with hematoxylin for 30 seconds. After bluing in tap water for 10 minutes, the slides were subjected to a gradient alcohol series for dehydration, cleared, and mounted. Digital imaging at 400× magnification facilitated the analysis of IL-6 and TNF-α protein expression within the colonic tissues.
Quantitative Real-Time PCR (qRT‒PCR)
We extracted total RNA from colonic tissues via a rapid extraction protocol using a Vazyme RNA extraction kit (RC112-01). The RNA was then reverse-transcribed to cDNA. By employing cDNA as a template and β-actin as an endogenous control, we performed amplification according to the instructions provided by the PCR amplification kit (Promega, a6001). The reactions were conducted in a total volume of 20 µL. Data quantification was performed utilizing the comparative 2−ΔΔCt method. The primer sequences utilized for amplification are detailed in Supplementary Table S1.
Western Blot Analysis
Total protein was harvested from the colonic tissues, and concentrations were ascertained using the BCA protein assay (Kaiji, KGP902). An aliquot of 25 μg of protein from each sample was loaded onto gels to ensure a uniform volume, mass, and concentration across wells. After a one-hour blocking step in 5% nonfat milk, the membranes were thoroughly washed with TBST, and incubated with antibodies at 4°C overnight. The following day, after washing with TBST, the membranes were incubated with the corresponding secondary antibodies at ambient temperature for one hour and washed three times with TBST. Detection of protein bands was carried out using an enhanced chemiluminescence (ECL) system. The antibody specifications used were as follows: β-actin (66009-1-1 g) served as the loading control and was used at a dilution of 1:10000; target proteins, including HIF-1α (Abcam, ab179483), TLR4 (Proteintech, 66350-1-1 g), phosphorylated NF-κB p65 (ser468) (Proteintech, 82335-1-RR), and NF-κB p65 (Proteintech, 66535-1-ig), were diluted to 1:1000.
Experimental Cells
Establishment, Grouping, and Treatment of the UC Cell Inflammatory Model
NCM460 cells (BNCC339288, China) were cultured in DMEM within a cell incubator set at 37°C and 5% CO2. Upon reaching an approximate cell confluence of 85%, the cells were passaged. Subsequently, the cells were divided into four distinct groups: the normal, model, BBR-treated, and inhibitor-treated groups. The latter three groups received 50 µg/mL LPS (L6529, USA)38 for 24 hours to induce an inflammatory response. Following this induction phase, the normal and model groups were maintained in a complete culture medium. The BBR group was treated with 10 µmol/L Berberine (02069–5g, China) >97% purity, and the inhibitor group was treated with 20 µM KC7F2 (HY-18777, USA).39
Monitoring of NCM460 Cells Following BBR Treatment
NCM460 cells in the logarithmic growth phase were seeded at a density of 3×105 cells per well in 6-well plates. To establish an inflammatory cell model, the cells were subjected to a 12-hour LPS challenge. Using optical microscopy, we documented the morphology of each cell group before and after BBR treatment at the 24-hour mark, enabling the assessment of treatment effects on cell conditions.
QRT-PCR Analysis
QRT-PCR was used to measure the mRNA expression levels of IL-6, TNF-α, IL-1β, TLR4, NF-κB, and HIF-1α. The sequences of the primers used are provided in Supplementary Table S2. The methodology and calculations were adapted from protocols established in mouse studies.
Protein Expression Analysis Through Western Blotting
Western blotting was used to measure the protein expression of TLR4, NF-κB, and HIF-1α. The detailed protocols, in addition to the data analysis procedures, were consistent with those developed for cell-based assays.
Statistical Analysis
The data were analyzed statistically using GraphPad Prism 8 software. Group comparisons were performed with one-way ANOVA. A P value of less than 0.05 was considered indicative of statistically significant differences.
Results
Network Pharmacology Predictive Outcomes
Identification of Potential Intervention Targets
Our study screened a set of 176 potential targets for BBR intervention and a comprehensive collection of the targets is outlined (Supplementary Table S3). After analyzing the GSE107499 dataset, we discerned 1193 differentially expressed genes DEGs (Figure 2A). The expression of key genes such as SLC25A33, TBPL2, and MSWIM3 was upregulated, whereas that of DUOX2, VNN1, and CEP112 was downregulated in UC tissue compared to healthy control tissue (Figure 2B). We retrieved 2918 disease-related targets from the GeneCards and Disgenet databases, identifying 119 targets intersecting BBR and UC (Figure 2C).
Construction and Analysis of the PPI Network
PPI network centered around the 119 intersecting targets, nodes displaying a higher degree highlighted their significance in the modulation of UC by BBR. Of particular interest were pivotal targets, such as TP53, HIF-1α, and TNF-α (Figure 3).
 |
Figure 3 PPI network of common targets of BBR against UC. |
GO and KEGG Enrichment Analysis
Enrichment analysis indicated that the therapeutic effects of BBR on UC predominantly pertain to Biological Processes (BPs) associated with cellular responses to chemical stress, variations in oxygen concentrations, and adaptation to hypoxia (Figure 4A). BBR’s impact on UC involves key cellular components (CCs), such as transcriptional regulatory complexes, lumens of secretory granules, and membrane microdomains (Figure 4B). Crucially, molecular functions (MFs), such as the binding activities of DNA-binding transcription factors, nuclear receptors, and transcriptional coregulators were also involved (Figure 4C). Finally, the investigation revealed several pathways that warrant focused consideration in the context of BBR’s therapeutic strategy for UC, including lipid and atherosclerosis pathways, the TNF signaling pathway, and the HIF-1 signaling pathway (Figure 4D).
 |
Figure 4 Results of GO and KEGG Analysis. (A) GO-Biological Process (BP). (B) GO-Cell Component (CC). (C) GO-Molecular Function (MF). (D) KEGG pathway analysis. |
Molecular Docking
Molecular docking was performed for seven pivotal targets with BBR and the corresponding inhibitors. The Protein Data Bank (PDB) identifiers for TLR4, HIF-1α, IL-6, TNF-α, IL-1β, NF-κB, and IL-10 are denoted as 7MLM, 5L9B, 1ALU, 7KPA, 5R86, 1IKN, and 1Y6M, respectively. The absolute values of the molecular docking interaction energies score Results are all greater than 5 kcal/mol (Supplementary Table S4). The interaction energetics score results suggest that BBR may interact with core proteins in the HIF pathway and further exert its effects. Notably, the effectiveness of TNF-α, HIF-1α, and TLR4 binding was particularly remarkable. We made the optimal 3D and 2D binding mode visualizations of BBR docking with HIF-1α (Figure 5 A1-A2), TLR4 (Figure 5 B1-B2), TNF-α (Figure 5 C1-C2), KC7F2 docking with HIF-1α (Figure 5 D1-D2), TLR4 (Figure 5 E1-E2), and TNF-α (Figure 5 F1-F2).
 |
Figure 5 Molecular Docking Mode Diagrams (A) BBR - HIF-1α. (B) BBR - TLR4. (C) BBR - TNF-α. (D) KC7F2 - HIF-1α. (E) KC7F2 - TLR4. (F) KC7F2 - TNF-α. |
Molecular Dynamics Simulation
Molecular dynamics simulations were used to delineate the dynamic interactions of the compound-target complexes, as depicted in the root-mean-square deviation (RMSD) plots (Figure 6). Across the simulation interval, interactions between protein structures and associated ligands—one of which included the compound of interest—were meticulously analyzed. Furthermore, the root-mean-square fluctuation (RMSF) profiles were normalized and graphed to capture fluctuations throughout the simulation’s timeline (Figure 7). The results confirmed the robust affinity and stability of the core targets and their respective compounds, validating the robustness of the screening process. A noteworthy aspect involved certain amino acids displaying marked variances in RMSD and RMSF representations, predominantly attributable to their pronounced flexibility within loop regions, whereas the remainder exhibited negligible conformational disturbances, corroborating the minor shifts in overall structural integrity.
 |
Figure 6 RMSD Charts During the Molecular Dynamics Simulation Process. (A) HIF-1α-BBR. (B) TLR4-BBR. (C) TNF-α-BBR. (D) HIF-α-KC7F2. (E) TLR4-KC7F2. (F) TNF-α-KC7F2. |
 |
Figure 7 RMSF Charts for the Molecular Dynamics Simulation of the Three Core Targets with Berberine and Inhibitors. (A) HIF-1α. (B) TLR4. (C) TNF-α. |
Validation Through Animal Experiments
Berberine Ameliorates DAI and Anal Lesions in DSS-Induced Mice
The experimental procedures for murine modeling and drug administration were employed in this study (Figure 8A). Observations made throughout the experiment indicated a pronounced improvement in mice treated with berberine after 7 days. In contrast to the model group—which exhibited lethargy, lackluster fur, and persistent diarrhea with hematochezia—the berberine-treated mice maintained robust conditions, with lustrous fur and feces akin to those of the control group. Both the DAI scores and alterations in body weight serve as poignant reflections of the inflammatory status of the mice, providing critical metrics for the assessment of the establishment of the UC model. Compared to those in the control group, significant exacerbation of DAI scores and a reduction in body weight were observed within the model group—a trend that was markedly reversed in mice treated with berberine and mesalazine, as evidenced by substantially lowered DAI scores and increased body weight (P < 0.01; Figure 8B and C). Notably, at the end of the treatment period, mice in the berberine group manifested neither anal redness nor bleeding, in stark contrast to the symptoms of the model group, thereby underscoring the potential of BBR in mitigating UC-associated rectal hemorrhage (Figure 8D).
Berberine Enhances Colonic Health, the CMDI, and the Splenic Index in DSS-Induced Mice
The progression of UC is known to impact the colonic length, CMDI, and splenic index in mice. After a 7-day therapeutic regimen, mice within the model group exhibited pronounced colonic inflammation and significant shortening of the colon when juxtaposed with those in the control group. In comparison, treatment with both berberine and mesalazine mitigated colonic inflammation and augmented colon length (P < 0.01; Figure 9A and B). The CMDI score offers an incisive assessment of intestinal mucosal integrity. While the model group of mice presented mucosal erosion, congestion, and edema, the berberine and mesalazine cohorts demonstrated intact mucosa with only minimal congestion and edema. This distinction was reflected quantitatively as a substantial decrease in CMDI scores (P < 0.05; Figure 9C and D). Furthermore, the splenic index is directly related to systemic inflammatory activity; a heightened index indicates increased inflammation.40 These findings imply that both berberine and mesalazine treatment regimens were efficacious at diminishing the splenic index, thereby attenuating the inflammatory response (P < 0.05; Figure 9E and F).
Berberine Mitigates Pathology in the Colon, Spleen, and Lungs of DSS-Induced Mice
To clarify the impact of berberine on pathological changes in a mouse model of UC, H&E staining was conducted on the colon, lung, and spleen tissues of the subjects. Analysis revealed extensive mucosal damage, crypt loss, glandular disorganization, and marked neutrophil infiltration in the model group. Treatment with berberine and mesalazine notably restored the integrity of the intestinal mucosa and crypts, resulting in a histological appearance nearly indistinguishable from that of the unaffected control group (Figure 10A).
 |
Figure 10 The pathological impact of berberine (400×). (A) Colon H&E. (B) Lung H&E. (C) Spleen H&E. |
H&E staining revealed the red pulp areas within the spleen, which are pivotal for the storage and release of erythrocytes and contribute to blood homeostasis. Conversely, the white pulp serves as a crucial immune system component. The distribution of red and white pulp is indicative of the inflammatory state within the organism.41 In the model mice, the blending of the medullary and red pulp sectors obscured their delineation. Berberine intervention led to the re-establishment of sharp segregation between these areas, closely resembling that of the control group (Figure 10B).
Drawing on the principles of traditional Chinese medicine, which underscore a significant functional interplay between the lung and colon, we posit that pathologies within the colon are likely to precipitate analogous changes within the lung.42 Correspondingly, the lung tissues of the model group exhibited increased alveolar wall thickness and intense inflammatory cell infiltration. Berberine treatment facilitated the reversion to normal alveolar structures and mitigated inflammatory activity (Figure 10C).
Berberine Diminishes Inflammatory Responses in DSS-Induced Mice
This investigation investigated the use of immunohistochemistry to track the expression and localization of the proinflammatory cytokines IL-6 and TNF-α in colonic tissues. Notably, the levels of these cytokines were elevated in the model group. After treatment with berberine and mesalazine, these inflammatory mediators were significantly downregulated (P < 0.05; Figure 11).
Berberine Attenuates TLR4/NF-κB/HIF-1α mRNA Expression in DSS-Induced Mice
This investigation investigated the influence of BBR on UC through the TLR4/NF-κB/HIF-1α pathway. QRT-PCR was used to quantify the mRNA expression levels of TLR4, NF-κB, and HIF-1α. BBR significantly reduced TLR4, NF-κB, and HIF-1α mRNA expression, exhibiting efficacy analogous to that of mesalazine (P < 0.05; Figure 12).
Berberine Reduces Protein Expression in the TLR4/NF-κB/HIF-1α Pathway in DSS-Induced Mice
Further analysis via Western blotting probed the effect of BBR on key proteins within the TLR4/NF-κB/HIF-1α pathway. The results demonstrated substantial suppression of TLR4, phosphorylated NF-κB, and HIF-1α protein expression in the presence of BBR compared to that in the untreated model group (P < 0.05; Figure 13). These findings suggest the therapeutic potential of BBR in UC through its regulatory effects on the TLR4/NF-κB/HIF-1α signaling axis.
Validation of Results Through Cellular Experiments
Restorative Effects of BBR on NCM460 Cells Post-LPS Challenge
Initially, the morphological assessment of the four cell groups revealed normalcy and viability (Figure 14A). Post-LPS challenge for 24 hours resulted in compromised cellular integrity, characterized by shrinkage, indistinct boundaries, and increased fragmentation (Figure 14B). In contrast, after treatment with BBR and a specific inhibitor, cellular morphology markedly improved, as indicated by the distinct contours, fewer fragments, and progressive normalization of cell conditions in the BBR-treated group compared to the LPS-treated model group (Figure 14C).
 |
Figure 14 Effects of BBR on cell status (200x). (A) Cell status before LPS intervention. (B) Cell status after LPS intervention. (C) Cell status after BBR treatment. |
Effect of Berberine on Inflammatory Cytokines and the mRNA Expression of TLR4, NF-κB, and HIF-1α in Cellular Models
Cell-based assays revealed that BBR significantly decreased the mRNA expression of the proinflammatory cytokines IL-6, IL-1β, and TNF-α (P<0.05, Figure 15A-C). To explore the molecular mechanism by which BBR affects UC, we analyzed the expression levels of TLR4, NF-κB, and HIF-1α. The analysis confirmed that BBR effectively downregulates the transcription of the genes integral to the TLR4/NF-κB/HIF-1α pathway (P<0.05, Figure 15D-F). Notably, in the inhibitor-treated group, KC7F2—a specific inhibitor of HIF-1α—reduced HIF-1α mRNA expression without altering the transcription of the upstream genes TLR4 and NF-κB.
Inhibition of Protein Expression by Berberine Along the TLR4/NF-κB/HIF-1α Pathway in a UC Cell Model
Western blotting provided insights into the protein-level effects of BBR in UC cellular models. Compared to those in the model group, the levels of TLR4, the ratio of phosphorylated to total NF-κB, and HIF-1α in the BBR treatment group were significantly lower (P<0.05; Figure 16). These findings indicate “the potential of BBR to attenuate the inflammatory response and ameliorate the hypoxic conditions associated with UC by targeting the TLR4/NF-κB/HIF-1α pathway”. The confirmation of the therapeutic impact of BBR was further supported by the observed reduction in HIF-1α expression in the inhibitor-treated group, which highlights the pivotal role of this protein in the disease mechanism and the efficacy of BBR in modulating this cellular pathway.
Molecular Mechanisms Underlying the Therapeutic Effect of BBR in UC
Integrated analyses utilizing pharmacological networking, coupled with empirical findings from in vivo and in vitro studies, have suggested that BBR may exert therapeutic effects on UC by modulating the TLR4/NK-NF-κB/HIF-1α signaling pathway. A schematic illustrating this molecular action depicts the interference of berberine: LPS engages the cell surface TLR4 receptor, culminating in the TLR4-LPS complex. This event initiates a cascade involving myeloid differentiation primary response 88 (MyD88) and TNF receptor-associated factor 6 (TRAF6), precipitating the phosphorylation and subsequent degradation of the NF-κB inhibitor (IκB). The resultant active NF-κB factor may then regulate the expression of various genes, either directly or indirectly, thereby modulating HIF-1α levels and influencing UC pathogenesis (Figure 17).
 |
Figure 17 Molecular Mechanism of BBR Treatment for Ulcerative Colitis via TLR4/NF-κB/HIF-1α Pathway. By Figdraw. |
Discussion
The intricate pathology of UC renders it a challenging disease to manage, if not recalcitrant to cure because severe cases necessitate colectomy. Moreover, the elongation of the disease trajectory enhances the risk of patient malignancy, jeopardizing patient well-being and survival.43 Clinical evidence suggests that berberine ameliorates UC by increasing the clinical response rate, facilitating mucosal reparative processes, and diminishing the Baron score.44 Corroborative basic research further revealed that berberine mitigates UC symptoms through mechanisms that include reducing intestinal mucosal damage and spleen indices, promoting autophagy, and suppressing lysozyme activity.45 Although naturally sourced, the anti-inflammatory properties of BBR position it as a substance of considerable therapeutic potential for UC management.46 However, a comprehensive understanding of the underlying mechanism of this phenomenon has not been achieved, providing the impetus for the present study.
In this investigation, network pharmacology was utilized to predict the primary targets of BBR in the management of UC. Analysis of the GEO clinical database revealed differentially expressed genes, including DUOX2 and SLC6A1447, which are implicated in the regulation of the HIF gene pathway. Notably, these genes may modulate the inflammatory responses of UC epithelial cells via the C/EBPβ-PAK6 axis, consistent with the findings of Brown and colleagues, who highlighted the relationship between mucosal hypoxia and inflammation severity in UC patients.
PPI network topological assessment has predicted BBR targets, such as TP53 and TNF-α, in UC treatment. These proteins are integral to a range of biological functions encompassing the hypoxic microenvironment of gastrointestinal epithelial cells, inflammation, and cell cycle governance.47,48 Furthermore, KEGG pathway analysis suggests that the HIF-1α pathway may associated with inflammatory and hypoxic response.49 These findings suggest that BBR may ameliorate UC by modulating the TLR4/NF-κB/HIF-1α signaling pathway. The results of molecular docking and molecular dynamics simulations suggest that BBR may have a favorable binding affinity with key proteins TLR4, NF-κB and HIF-1α.
However, despite these promising indications, network pharmacology offers only a theoretical prediction of the therapeutic potential of BBR via the TLR4/NF-κB/HIF-1α pathway in UC, and further empirical validation is lacking.50 All computational models are mere suggestions and no proof for anything.The computational models results are merely called a “hypothesis” and need further validation through experimentation.Thus, experimental mouse models and cellular assays were employed to verify these postulations. The administration of DSS, a colitogenic agent commonly utilized to simulate UC in murine models, induces substantial destruction of the intestinal epithelial architecture and inflammatory influx, typified by symptoms such as diarrhea, rectal bleeding, weight decrement, and reduced colon length.51 Our in vivo findings revealed that BBR treatment markedly counters these adverse effects, improving anal inflammation and bleeding, as indicated by DAI scores; weight; colon length; and spleen size approaching those of healthy controls. Additionally, UC pathogenesis involves exacerbated inflammatory reactions and immune dysregulation, paralleled by histological distortions. H&E staining has shown that BBR notably mitigates intestinal, splenic, and pulmonary pathological defects, restoring anatomical normalcy. Given the contributory role of proinflammatory cytokines in UC pathogenesis, the therapeutic objective of modulating these mediators is fundamental. Our use of qRT-PCR to assess the anti-inflammatory efficacy of BBR yielded encouraging data indicating decreases in proinflammatory cytokines, such as IL-6, TNF-α, and IL-1β. This study’s findings not only deepen the understanding of BBR’s therapeutic mechanisms but also fortify its potential as an intervention for UC.
Following the assessment of the anti-inflammatory efficacy of berberine in a murine model of UC, this study investigated the molecular mechanisms underlying the TLR4/NF-κB/HIF-1α signaling pathway, as hypothesized by network pharmacological analyses. TLR4, a pivotal inflammatory mediator ubiquitously present in numerous immune and endothelial cells, is implicated in the pathogenesis of intestinal inflammation and apoptosis.18 In the setting of UC, enhanced expression of TLR4 and its downstream effector NF-κB precipitates a dysregulation of immune functions; therefore, targeting the TLR4/NF-κB axis represents a viable therapeutic strategy for ameliorating UC.52–54 The NF-κB signaling cascade is intimately associated with proinflammatory mediators. Notably IL-6 and TNF-α—cytokines increase in conjunction with the progression of UC. Data from mechanistic inquiries have demonstrated that berberine attenuates the mRNA and protein expression of both TLR4 and NF-κB. Moreover, HIF-1α induction is disrupted in the absence of NF-κB activity, even under sustained hypoxic conditions, suggesting that HIF-1α transcription is predominantly governed by NF-κB, thereby revealing the integral TLR4/NF-κB/HIF-1α signaling pathway. Crucially, HIF-1α—a regulator of macrophage function—binds to hypoxic response elements in concert with HIF-1β, playing a central role in facilitating the recovery and replication of inflamed epithelial cells. A heightened HIF-1α expression corresponds to an exacerbation of the inflammatory milieu.19 Thus, its inhibition emerges as a therapeutic axis to curb UC.55–57 In vivo experiments have corroborated these findings, indicating that berberine substantially diminishes both the mRNA and protein expression of HIF-1α.
In vitro, BBR was administered to LPS-stimulated UC cell models, and the cellular morphology was evaluated via microscopy. Cells treated with BBR presented enhanced structural integrity and reduced fragmentation, indicating the potential for organelle rejuvenation and cytoprotective effects. Concordance was found between the qRT-PCR and Western blot analyses and in vivo outcomes, with BBR eliciting a decrease in the mRNA and protein expression of TLR4, NF-κB, and HIF-1α. The use of the HIF-1α inhibitor KC7F2 in cellular assays revealed that while the expression of TLR4 and NF-κB was unaltered, marked suppression of HIF-1α was observed, further confirming the role of BBR in modulating the hypoxic response via the TLR4/NF-κB/HIF-1α pathway to mitigate UC inflammation. This finding supports the accuracy of the predictions made by our network pharmacology approach.
In the present investigation, we confined our predictive analysis within the scope of specific databases and pharmacological repositories for network pharmacology, a limitation that may introduce biases in our findings. Future endeavors should aim to incorporate a broader array of data sources and validate findings through a more extensive set of experimental procedures to bolster the reliability of the predictions. Moreover, a concerted effort to assess the efficacy of network pharmacology in medicinal discovery and design, combined with empirical and clinical research, constitutes a promising avenue of inquiry.
Conclusion
This study utilizes bioinformatics and in vivo and in vitro experimental validation methods to preliminarily suggest that BBR may treat UC by alleviating hypoxic responses through inhibition of the TLR4/NF-κB/HIF-1α signaling pathway. In summary, this study provides meaningful insights into the molecular mechanisms underlying berberine-assisted treatment of UC.
Abbreviations
UC, ulcerative colitis; BBR, berberine; IBD, inflammatory bowel disease; DAI, disease activity index TNF-α, tumor necrosis factor alpha; IL-6, interleukin 6; IL-1β, interleukin 1β; IL-10, interleukin 10; qRT‒PCR, Quantitative Real-Time PCR; 5-ASA,5-Amino salicylic acid; MPO, Manufacturing Production Order; MD Molecular docking; MDS, Molecular dynamics simulations; GO, Gene Ontology; NF-κB, RMSD, Root Mean Square Displacement; RMSF, Root Mean Square Fluctuation; LPS, Lipopolysaccharide; BP, Biological Process; CCK-8, Cell Counting Kit-8; IκB, Inhibitor of NF-κB; C/EBPβ, CCAAT-enhancer-binding protein β; Th17, T helper cell 17; IFN-γ, Interferon-gamma; CC, Cell Component.
Ethics Statement
This study involves the use of the GEO database, as it is a public database for human data. Ethical approval for the research was granted by the Ethics Committee of the Beijing Institute of Traditional Chinese Medicine (approval number: BJTCM-M-2023-10-17). Ethical approval for animal experimentation was also granted (approval number: BJTCM-M-2023-11-07). GEO belong to public databases. The patients involved in the database have obtained ethical approval. Users can download relevant data for free for research and publish relevant articles. Our study is based on open source data, so there are no ethical issues.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work. Jilei Li and Wenchao Dan are the co-first authors, Shengsheng Zhang are the corresponding author.
Funding
This study was supported by National Natural Science Foundation of China (82174304).
Disclosure
The authors declare that there are no conflicts of interest in this work.
References
1. Ungaro R, Mehandru S, Allen PB, Peyrin-Biroulet L, Colombel JF. Ulcerative colitis. Lancet. 2017;389(10080):1756–1770.
2. Mak WY, Zhao M, Ng SC, Burisch J. The epidemiology of inflammatory bowel disease: east meets west. J Gastroenterol Hepatol. 2020;35(3):380–389.
3. Shah SC, Itzkowitz SH. Colorectal Cancer in Inflammatory Bowel Disease: mechanisms and Management. Gastroenterology. 2022;162(3):715–730e713.
4. Paik J. Ozanimod: a Review in Ulcerative Colitis. Drugs. 2022;82(12):1303–1313.
5. Hanzel J, Hulshoff MS, Grootjans J, D’Haens G. Emerging therapies for ulcerative colitis. Expert Rev Clin Immunol. 2022;18(5):513–524.
6. Liu Y, Li BG, Su YH, et al. Potential activity of Traditional Chinese Medicine against Ulcerative colitis: a review. J Ethnopharmacol. 2022;289:115084.
7. Zhu C, Li K, Peng XX, et al. Berberine a traditional Chinese drug repurposing: its actions in inflammation-associated ulcerative colitis and cancer therapy. Front Immunol. 2022;13:1083788.
8. Wu J, Luo Y, Shen Y, et al. Integrated Metabonomics and Network Pharmacology to Reveal the Action Mechanism Effect of Shaoyao Decoction on Ulcerative Colitis. Drug Des Devel Ther. 2022;16:3739–3776.
9. Luo Y, Wu J, Liu Y, et al. Metabolomics Study of Shaoyao Plants Decoction on the Proximal and Distal Colon in Mice with Dextran Sulfate Sodium-Induced Colitis by UPLC-Q-TOF-MS. Drug Des Devel Ther. 2022;16:4343–4364.
10. Lu QK, Fu YF, Li H. Berberine and its derivatives represent as the promising therapeutic agents for inflammatory disorders. Pharmacol Rep. 2022;74(2):297–309.
11. Khoshandam A, Imenshahidi M, Hosseinzadeh H. Pharmacokinetic of berberine, the main constituent of Berberis vulgaris L.: a comprehensive review. Phytother Res. 2022;36(11):4063–4079.
12. Tang X, Yang M, Gu Y, Jiang L, Du Y, Liu J. Orally Deliverable Dual-Targeted Pellets for the Synergistic Treatment of Ulcerative Colitis. Drug Des Devel Ther. 2021;15:4105–4123.
13. Cooper SF, Mockle JA, Beliveau J. Alkaloids of Coptis groenlandica. Planta Med. 1970;19(1):23–29.
14. Xu Y, Huang J, Fan Y, et al. Macrophage-Targeted Berberine-Loaded beta-Glucan Nanoparticles Enhance the Treatment of Ulcerative Colitis. Int J Nanomed. 2022;17:5303–5314.
15. Dong YL, Fan H, Zhang Z, et al. Berberine ameliorates DSS-induced intestinal mucosal barrier dysfunction through microbiota-dependence and Wnt/β-catenin pathway. Int J Biol Sci. 2022;18(4):1381–1397.
16. Li H, Fan C, Lu HM, et al. Protective role of berberine on ulcerative colitis through modulating enteric glial cells-intestinal epithelial cells-immune cells interactions. Acta Pharmacol Sin B. 2020;10(3):447–461.
17. Zhou Y, Yang S, Guo J, et al. In Vivo Imaging of Hypoxia Associated with Inflammatory Bowel Disease by a Cytoplasmic Protein-Powered Fluorescence Cascade Amplifier. Anal Chem. 2020;92(8):5787–5794.
18. Dai W, Long L, Wang X, Li S, Xu H. Phytochemicals targeting Toll-like receptors 4 (TLR4) in inflammatory bowel disease. Chin Med. 2022;17(1):53.
19. Dvornikova KA, Platonova ON, Bystrova EY. Hypoxia and Intestinal Inflammation: common Molecular Mechanisms and Signaling Pathways. Int J Mol Sci. 2023;24(3).
20. Vidal-Limon A, Aguilar-Toala JE, Liceaga AM. Integration of Molecular Docking Analysis and Molecular Dynamics Simulations for Studying Food Proteins and Bioactive Peptides. J Agric Food Chem. 2022;70(4):934–943.
21. Gallo K, Goede A, Preissner R, Gohlke BO. SuperPred 3.0: drug classification and target prediction-A machine learning approach. Nucleic Acids Res. 2022;50(W1):W726–W731.
22. Liu X, Ouyang S, Yu B, et al. PharmMapper server: a web server for potential drug target identification using pharmacophore mapping approach. Nucleic Acids Res. 2010;38:W609–W614.
23. Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47(W1):W357–W364.
24. Fang S, Dong L, Liu L, et al. HERB: a high-throughput experiment- and reference-guided database of traditional Chinese medicine. Nucleic Acids Res. 2021;49(D1):D1197–D1206.
25. Xu HY, Zhang YQ, Liu ZM, et al. ETCM: an encyclopaedia of traditional Chinese medicine. Nucleic Acids Res. 2019;47(D1).
26. Kuhn M, Szklarczyk D, Franceschini A, et al. STITCH 2: an interaction network database for small molecules and proteins. Nucleic Acids Res. 2010;38(Database issue):D552–556.
27. Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13.
28. Wishart DS, Feunang YD, Guo AC, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–D1082.
29. UniProt C. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49(D1):D480–D489.
30. Szklarczyk D, Gable AL, Nastou KC, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605–D612.
31. Goodsell DS, Zardecki C, Di Costanzo L, et al. RCSB Protein Data Bank: enabling biomedical research and drug discovery. Protein Sci. 2020;29(1):52–65.
32. O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: an open chemical toolbox. J Cheminform. 2011;3:33.
33. Zhang Y, Sanner MF. AutoDock CrankPep: combining folding and docking to predict protein-peptide complexes. Bioinformatics. 2019;35(24):5121–5127.
34. Vilar S, Cozza G, Moro S. Medicinal chemistry and the molecular operating environment (MOE): application of QSAR and molecular docking to drug discovery. Curr Top Med Chem. 2008;8(18):1555–1572.
35. Hsin KY, Ghosh S, Kitano H. Combining machine learning systems and multiple docking simulation packages to improve docking prediction reliability for network pharmacology. PLoS One. 2013;8(12).
36. Cooper HS, Murthy SN, Shah RS, Sedergran DJ. Clinicopathologic study of dextran sulfate sodium experimental murine colitis. Lab Invest. 1993;69(2):238–249.
37. Bell CJ, Gall DG, Wallace JL. Disruption of colonic electrolyte transport in experimental colitis. Am J Physiol. 1995;268(4):G622–630.
38. Wu D, Zhang Y, Zou B, Lu Y, Cao H. Shaoyao decoction alleviates TNBS-induced ulcerative colitis by decreasing inflammation and balancing the homeostasis of Th17/Treg cells. BMC Complement Med Ther. 2023;23(1):424.
39. Narita T, Yin S, Gelin CF, et al. Identification of a novel small molecule HIF-1alpha translation inhibitor. Clin Cancer Res. 2009;15(19):6128–6136.
40. Sun L, Nie X, Lu W, et al. Mucus-Penetrating Alginate-Chitosan Nanoparticles Loaded with Berberine Hydrochloride for Oral Delivery to the Inflammation Site of Ulcerative Colitis. AAPS Pharm Sci Tech. 2022;23(6):179.
41. Orazi A, Arber DA. Pathology of the spleen: introduction. Semin Diagn Pathol. 2021;38(2):111.
42. Xutian S, Zhang J, Louise W. New exploration and understanding of traditional Chinese medicine. Am J Chin Med. 2009;37(3):411–426.
43. Wanders LK, Dekker E, Pullens B, Bassett P, Travis SP, East JE. Cancer risk after resection of polypoid dysplasia in patients with longstanding ulcerative colitis: a meta-analysis. Clin Gastroenterol Hepatol. 2014;12(5):756–764.
44. Xu L, Zhang Y, Xue X, et al. A Phase I Trial of Berberine in Chinese with Ulcerative Colitis. Cancer Prev Res (Phila). 2020;13(1):117–126.
45. Xu X, Li W, Yu Z, et al. Berberine Ameliorates Dextran Sulfate Sodium-Induced Ulcerative Colitis and Inhibits the Secretion of Gut Lysozyme via Promoting Autophagy. Metabolites. 2022;12(8).
46. Izadparast F, Riahi-Zajani B, Yarmohammadi F, Hayes AW, Karimi G. Protective effect of berberine against LPS-induced injury in the intestine: a review. Cell Cycle. 2022;21(22):2365–2378.
47. Escudero-Hernandez C, van Beelen Granlund A, Bruland T, et al. Transcriptomic Profiling of Collagenous Colitis Identifies Hallmarks of Nondestructive Inflammatory Bowel Disease. Cell Mol Gastroenterol Hepatol. 2021;12(2):665–687.
48. Chen Y, Yan W, Chen Y, et al. SLC6A14 facilitates epithelial cell ferroptosis via the C/EBPbeta-PAK6 axis in ulcerative colitis. Cell Mol Life Sci. 2022;79(11):563.
49. Brown E, Rowan C, Strowitzki MJ, et al. Mucosal inflammation downregulates PHD1 expression promoting a barrier-protective HIF-1alpha response in ulcerative colitis patients. FASEB J. 2020;34(3):3732–3742.
50. Yuan Z, Pan Y, Leng T, et al. Progress and Prospects of Research Ideas and Methods in the Network Pharmacology of Traditional Chinese Medicine. J Pharm Pharm Sci. 2022;25:218–226.
51. Li H, Wang Y, Shao S, et al. Rabdosia serra alleviates dextran sulfate sodium salt-induced colitis in mice through anti-inflammation, regulating Th17/Treg balance, maintaining intestinal barrier integrity, and modulating gut microbiota. J Pharm Anal. 2022;12(6):824–838.
52. Tam JSY, Coller JK, Hughes PA, Prestidge CA, Bowen JM. Toll-like receptor 4 (TLR4) antagonists as potential therapeutics for intestinal inflammation. Indian J Gastroenterol. 2021;40(1):5–21.
53. Yousefi-Manesh H, Dejban P, Mumtaz F, et al. Risperidone attenuates acetic acid-induced colitis in rats through inhibition of TLR4/NF-kB signaling pathway. Immunopharm Immunot. 2020;42(5):464–472.
54. Rashidian A, Muhammadnejad A, Dehpour AR, et al. Atorvastatin attenuates TNBS-induced rat colitis: the involvement of the TLR4/NF-kB signaling pathway. Inflammopharmacology. 2016;24(2–3).
55. Yin J, Ren Y, Yang K, et al. The role of hypoxia-inducible factor 1-alpha in inflammatory bowel disease. Cell Biol Int. 2022;46(1):46–51.
56. Li MY, Luo HJ, Wu X, et al. Anti-Inflammatory Effects of Huangqin Decoction on Dextran Sulfate Sodium-Induced Ulcerative Colitis in Mice Through Regulation of the Gut Microbiota and Suppression of the Ras-PI3K-Akt-HIF-1α and NF-κB Pathways. Front Pharmacol. 2020;10.
57. Zhuang H, Lv Q, Zhong C, et al. Tiliroside Ameliorates Ulcerative Colitis by Restoring the M1/M2 Macrophage Balance via the HIF-1alpha/glycolysis Pathway. Front Immunol. 2021;12:649463.
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.








