Back to Journals » Journal of Inflammation Research » Volume 17
Gut Microbiota Disorder Contributes to the Production of IL-17A That Exerts Chemotaxis via Binding to IL-17RA in Endometriosis
Authors Li Y, Zhou Z, Liang X , Ding J, He Y, Sun S, Cheng W, Ni Z, Yu C
Received 10 January 2024
Accepted for publication 18 June 2024
Published 1 July 2024 Volume 2024:17 Pages 4199—4217
DOI https://doi.org/10.2147/JIR.S458928
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Yangshuo Li,1,* Zhihao Zhou,1,2,* Xiaolan Liang,1,* Jie Ding,1 Yalun He,1 Shuai Sun,1 Wen Cheng,1 Zhexin Ni,1,3 Chaoqin Yu1
1Department of Traditional Chinese Gynecology, the First Affiliated Hospital of Naval Military Medical University (Changhai Hospital), Shanghai, People’s Republic of China; 2Traditional Chinese Medicine Department, No. 929 Hospital, Naval Medical University, Shanghai, People’s Republic of China; 3Department of Pharmaceutical Sciences, Beijing Institute of Radiation Medicine, Beijing, 100850, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zhexin Ni, Department of Pharmaceutical Sciences, Beijing Institute of Radiation Medicine, 27 Taiping Road, Beijing, 100850, People’s Republic of China, Email [email protected] Chaoqin Yu, Department of Traditional Chinese Gynecology, the First Affiliated Hospital of Naval Military Medical University (Changhai Hospital), 168 Changhai Road, Shanghai, 200433, People’s Republic of China, Email [email protected]
Introduction: Endometriosis (EM) is a chronic estrogen-dependent condition characterized by the growth of endometrial-like tissue outside the uterus, posing a significant burden on reproductive-aged women. Previous research has shown a correlation between gut microbiota dysbiosis and interleukin-17A (IL-17A) in EM patients. IL-17A, a promising immunomodulatory molecule, exerts dual roles in human physiology, driving inflammatory diseases. However, the functions and origins of IL-17A in EM remain poorly characterized.
Methods: Single-cell data analysis was employed to characterize IL-17A activity in EM lesions. Fecal microbiota transplantation was conducted to explore the impact of gut microbiota on EM. Gut microbiota and bile acid metabolism were assessed via 16S rRNA sequencing and targeted metabolomics. Th17 cell proportions were measured using flow cytometry.
Results: High expression of IL-17 receptor A (IL-17RA) was observed in myeloid cell subpopulations within EM lesions and may be involved in the migration and recruitment of inflammatory cells in lesions. Elevated IL-17A levels were further validated in peritoneal and follicular fluids of EM patients. Dysregulated bile acid levels, particularly elevated chenodeoxycholic acid (CDCA) and ursodeoxycholic acid (UDCA), were found in the gut and peritoneal fluid of EM mouse models. Additional CDCA administration reduced EM lesions and modulated Th17 cell proportions, while UDCA showed no significant effects.
Discussion: Our findings shed light on the origins and functions of IL-17A in EM, implicating its involvement in lesion migration and recruitment. Dysregulated bile acid metabolism may contribute to EM pathogenesis, with CDCA exhibiting therapeutic potential.
Keywords: endometriosis, single-cell sequencing, interleukin-17, gut microbiota, bile acids, myeloid cell
Graphical Abstract:
Introduction
Endometriosis (EM) is an estrogen-dependent disease characterized by the presence of functional endometrial glands and stroma outside the uterus, most commonly found in the ovaries and pelvic peritoneum.1 It is a prevalent gynecological disorder, with reported incidence rates ranging from 2% to 10% in the general female population and up to 50% among infertile women.2 Several hypotheses exist regarding the pathogenesis of EM, with the theory of retrograde menstruation being the most widely accepted.3 However, despite the majority of women of reproductive age experiencing retrograde menstruation, EM only manifests in 10–15% of cases, suggesting the existence of other mechanisms contributing to its development.
Our previous research has demonstrated dysbiosis in the intestinal microbiota of EM patients, with dysregulated intestinal microbiota potentially contributing to both intestinal barrier damage and the progression of EM.4,5 However, the direct impact of the intestinal microbiota and microbiota-derived metabolites on the progression of EM remains largely unknown. Numerous studies have indicated subtle connections between the intestines and other organs, such as the gut-brain axis and gut-lung axis.6–8 This interplay may involve various pathways through which the microbiota interacts with distant organs, including the immune system, metabolic products, and the enteric nervous system, implicating microbiota-derived metabolites such as bile acids, short-chain fatty acids, and branched-chain amino acids.9–12
Moreover, emerging evidence suggests that bile acid metabolites in the gut regulate the balance between interleukin-17A (IL-17A)-producing helper T cells (Th17 cells) and regulatory T cells (Treg cells) to modulate host immune responses.13,14 IL-17 cytokines, produced by Th17 cells, possess diverse biological functions, serving to protect against infections but also potentially contributing to infections and autoimmune diseases.15 Among the IL-17 family members, IL-17A exerts significant immunological effects by binding to IL-17RA and IL-17RC.15 Additionally, gut Th17 cells play a dual role, not only serving as essential mediators of intestinal epithelial barrier integrity but also potentially participating in inducing intestinal inflammation.16 Several studies have underscored the association of intestinal Th17 populations with driving extraintestinal autoimmune diseases.17,18 These findings suggest that dysbiosis of the intestinal microbiota may further induce dysregulation of intestinal immune Th17 cell populations via alterations in bile acid metabolism and may be associated with extraintestinal diseases. Dysbiosis of the intestinal microbiota and bile acid metabolism profiles have been observed in EM, yet whether intestinal Th17 cell populations are dysregulated in EM remains unknown.
Recent studies have demonstrated a significant elevation of IL-17A levels in peripheral blood and peritoneal fluid of EM patients, particularly in the early stages, which may be linked to disease progression.19,20 However, the specific sources of IL-17A in the peritoneal fluid or lesions of EM patients and their downstream mechanisms remain to be elucidated. Additionally, some in vitro studies have identified expression of IL17RA and IL17RC in endometrial stromal cells,21,22 albeit with limitations, as immune cells can infiltrate both the endometrium and EM lesions.
Single-cell RNA sequencing (scRNA-seq) enables us to unravel the onset and progression of diseases at the level of individual cells, providing insights into the heterogeneity of gene expression within cell populations.23 Recent studies utilizing scRNA-seq have revealed the presence of diverse cell types within EM lesions, including stromal cells, epithelial cells, lymphocytes, myeloid cells, and endothelial cells.24 Moreover, different types of EM lesions exhibit cellular-level heterogeneity,24,25 underscoring the necessity of understanding the dynamic interactions between cells to elucidate the complex pathogenesis of EM.
Therefore, in this study, we identified high expression of IL-17RA in bone marrow cells within EM lesions at the single-cell level, suggesting its potential role as a target for IL-17A binding and exploring its downstream biological effects. Furthermore, we investigated whether the elevated levels of IL-17A in EM may stem from dysregulation of certain bile acid metabolites following dysbiosis of the intestinal microbiota.
Materials and Methods
Patient Sample Collection
All patients’ peritoneal fluid samples were obtained from the First Affiliated Hospital of Naval Medical University and the Ninth People’s Hospital of Shanghai. We recruited 15 women with infertility of EM and 15 women who underwent assisted reproduction due to the male partner’s reasons, etc., and some of the patients’ follicular fluids were collected at the time of oocyte retrieval for the study. The inclusion criteria for EM used in this study were patients diagnosed with EM by histology and without comorbid adenomyosis, uterine leiomyoma or polycystic ovary syndrome (PCOS) (age < 48 years). The main inclusion criteria for the control group were women undergoing assisted reproduction due to male infertility in their partners. The diagnostic criteria for male infertility were based on the World Health Organization Standardized Tests and Diagnostic Manual for Male Infertility.
In addition, 36 additional women who underwent laparoscopic surgery for ovarian endometriosis, uterine leiomyoma, or infertility were recruited. Patients who had received medications/hormonal supplements within the last 3 months or were diagnosed with malignancy or pelvic inflammation were excluded from recruitment. Among them, 24 cases of EM peritoneal fluid and 12 cases of non-EM peritoneal fluid were collected. The experiment was approved by the Ethics Committee of the First Affiliated Hospital of Naval Medical University (CHEC2019-100) and was conducted by the Declaration of Helsinki. All patients have signed informed consent forms for collecting clinical information.
ELISA
Cytokine IL-17A levels in peritoneal fluid and follicular fluid were measured using appropriate enzyme-linked immunosorbent assay (ELISA) kits (MultiSciences) according to the manufacturer’s instructions.
Animal Model
Seven-week-old female C57BL mice were purchased from Beijing Viton Lihua Laboratory Animal Technology Co. After one week of adaption, the mice were randomly divided into the control group (CON) and EMS model group (EM). They were housed in an SPF-grade environment in the Animal Center of Changhai Hospital in Shanghai, with 12 hours of light and 12 hours of darkness. The welfare of experimental animals is guided by Changhai Hospital Guidelines for Ethical Review of Experimental Animal Welfare. All experimental manipulations were approved by the Animal Management and Use Committee of Changhai Hospital in Shanghai.
The construction of the EMS mouse model was described in a previous study.4 Briefly, donor mice were injected subcutaneously with 0.2 μg/g estrogen solution on days 1, 4, and 7, and endometrial fragments were transplanted on day 8. Donor mice were anesthetized with ether, euthanized by neck dissection, and then immersed in 75% ethanol for 3 min. The uterus was obtained by dissecting and cutting along the longitudinal axis of the uterus. The endometrial layer was peeled off and cut into 1 mm × 1 mm × 1 mm pieces. The endometrial fragments were aspirated with a syringe, and donor and recipient mice were implanted intraperitoneally in a 1:2 ratio. The estrogen solution was injected subcutaneously on days 7 and 14 after transplantation. The endometrial transplantation procedures were performed under sterile conditions. The control group was injected intraperitoneally with saline containing adipose tissue under the same conditions. After different interventions were given, EM ectopic tissue and peritoneal fluid were collected for subsequent analysis. In performing the focal tissue assessment, it was conducted by an experimenter who was not aware of the grouping.
Fecal Microbiota Transplantation (FMT)
To investigate whether gut microbiota disorder promotes the progression of EM, we conducted FMT experiments.26 Briefly, fecal samples were processed under anaerobic conditions. Each donor mouse was placed in a clean plastic cage without food or bedding at the same time each day and observed until defecation (usually 5–10 min). Fecal material was then immediately transferred to sterile microcentrifuge tubes and stored at −80°C. For recipient mice, 0.2 mL of an antibiotic cocktail (vancomycin 0.5 g/L, penicillin 1 g/L, metronidazole 1 g/L, neomycin 1 g/L, Yuanye Bio Co.) was intragastric administrated daily for one week after completion of modeling. Subsequently, a certain amount of feces was extracted daily. Twenty milligrams of feces was mixed with 1 mL of sterile saline and shaken, and the supernatant was collected after centrifugation. Subsequently, the fecal suspensions of mice in EM and CON groups were intervened by intragastric administration in FMT-CON and FMT-EM groups for 2 weeks.
Assessment of Intestinal Permeability
Assessment of intestinal permeability is a method used to assess whether the intestinal barrier is compromised, aiding in our understanding of the mouse intestinal condition. As previously mentioned, with slight modifications.27 Mice were fasted in a clean cage without food or water. After 4 hours, mice were intragastric administrated with 0.6 mg/g 4 kDa fluorescein isothiocyanate dextran (Sigma), and after an additional 4 hours, blood was collected through orbital blood sampling. Donor mice were then euthanized via cervical dislocation. The collected blood was centrifuged at 2000 × g for 5 minutes, and the supernatant was extracted. Fluorescence intensity in the serum was measured by spectrophotometry (485nm/528nm).
Extraction of Intestinal Lamina Propria Cells
The lamina propria of the intestine is the richest reservoir of resident immune cells in the gut, which most accurately reflects the status of the intestinal immune microenvironment.28 Therefore, we extracted lamina propria cells from the intestines of mice to assess the proportion of intestinal Th17 cells in EM model mice. Mouse intestinal lamina propria immune cells were obtained as previously described29 but with slight modifications. The colon was placed on a petri dish containing an appropriate amount of pre-cooled 1× PBS buffer, fat on the intestinal wall was stripped, the intestine was cut longitudinally along the mesentery, and the contents of the intestinal lumen were washed with PBS buffer 3 to 4 times. Appropriately sized intestinal segments (1.5−2 cm) were first placed in 10 mL of pre-digestive solution 1 (containing 0.01 mL dithiothreitol (DTT, 1 mol/L) and 0.1 mL HEPES (1 mol/L) per 10 mL of PBS) and shaken horizontally for 10 min at 37 °C at 220 r/min, and then manually oscillated vigorously up and down for 1 min. Subsequently, the intestinal segments were placed in 10 mL of pre-digestive solution 2 (0.625 mL of EDTA (0.5 mol/L) and 0.1 mL of HEPES (1 mol/L) per 10 mL of PBS) and shaken horizontally at 37 °C for 10 min at 220 r/min, and then manually shaken vigorously up and down for 1 min, repeating the procedure. Pre-digestive tissues were washed twice with PBS and placed in 5 mL of RPMI-1640 medium containing 10% serum and then transferred to a 6-well plate containing 3−5 mL of digestion solution (RPMI-1640 with 10% FBS containing collagenase VIII (0.2−0.4 units/mL) and DNase I (0.15 mg/mL), mixed well and used, 3−5 mL per sample), and cut to less than 0.5 cm with sterile surgical forceps and scissors, last placed in a 37 °C and 5% CO2 cell culture incubator for 90 min. After digestion, the intestine segment was transferred to a 15 mL centrifuge tube, terminated digestion by adding 7 mL of PBS, manually vibrating vigorously for 2 min, next filtered the cells into a 50 mL centrifuge tube using a 70 μm filter membrane and volume to 35 mL with PBS, finally centrifuged at 1000 × g for 10 min at 4 °C. The cell precipitate was resuspended in the appropriate volume of FBS and used for flow cytometry analysis.
Flow Cytometry
Flow cytometry was used to detect changes in the proportion of specific immune cells. Cell suspensions were prepared as described above, blocked with mouse Fc block (BD Pharmingen), and stained with fixable viability dye (FVS510, BD Pharmingen) and surface antibodies (CD3-FITC, CD4-BB700, BD Pharmingen) in PBS 0.5% bovine serum albumin at corresponding concentrations. For the intracellular cytokine assay (IL-17a-AF647, FOPX3-PE-CY7, BD Pharmingen), cells were incubated with Leukocyte Activation Cocktail and BD GolgiPlug (BD Pharmingen) in complete RPMI medium at 37 °C for 4 hours. After surface staining, cells were fixed and permeabilized with the Transcription Factor Buffer Set (BD Pharmingen) at 4 °C overnight. Staining was performed using the relevant anti-cytokine antibodies in BD Perm/Wash buffer at a concentration of 1:100 for 1 hour at 4 °C. Flow cytometry was performed using CytoFLEX (Beckman Coulter, USA), and data were analyzed with FlowJo software. The gating procedure is shown in Figure S1.
Hematoxylin-Eosin Stain (H&E)
The 4 μ m thick tissue sections were dewaxed and rehydrated in ethanol and water. The slides were then stained with hematoxylin (Biosci, China) for 5 min and with eosin (Biosci, China) for 2 min.
Western Blotting
Mouse colon segments were washed 2–3 times with cold PBS and then cut into small pieces and placed in homogenization tubes. Lysis reagent with 10 times of tissue volume (protease inhibitor was added within minutes before use) and 1−2 small 4mm magnetic beads were added to a tissue grinder at 60Hz for 60s. The sample tube with homogenization completed was taken out and ice bath for 10min, which can be shaken appropriately to make sure that the tissue is completely lysed. The total protein solution that was supernatant was collected by centrifugating at 12,000rpm for 10 minutes. The protein concentration was measured by the BCA Protein Concentration Assay Kit (Beyotime, China). Using pre-stained protein molecular weight standards (SD0023/SD0024) as protein kDa ladder reference. After treatment on denaturation of protein, 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis was performed and then transferred to polyvinylidene difluoride membrane (Millipore). The proteins were further incubated with Claudin 1 primary antibody (1:1000, ab15098, Abcam, USA) and then with horseradish peroxidase-conjugated secondary antibody (1:20,000, SD0039, Simuwubio, China). Capture images using a gel imager (Tanon 4600) with the parameter set to automatic exposure. ImageJ software processing system was used to analyze the optical density values of the target bands.
Targeted Bile Acid Metabolomics
To detect whether there is dysregulation in the bile acid metabolism profile in the peritoneal fluid of EM model mice, we employed targeted bile acid metabolomics. The peritoneal fluid from the mice was collected and then centrifuged at 4 degrees 3000rpm for 10 minutes, dispensed in centrifuge tubes, quick-freeze in liquid nitrogen for 10 minutes, and stored in a −80 refrigerator. The 33 bile acid standards were weighed accurately to prepare a linear master mix at a concentration of 25 μg/mL and then were diluted with methanol to obtain a series of working solutions at concentrations of 25,000, 15,000, 5000, 2500, 500, 250, 50, 25, 15, 5, 2.5, and 1.5 ng/mL. The internal standard solution (IS) was obtained through certain concentrations of GCA-d4, LCA-d4, CDCA-d4, UDCA-d4, CA-d4, and GCDCA-d4 solutions mixing. The master and working solutions of linear, internal standard, and mass control were stored in a −20°C refrigerator. The diluted sample (see the final report for the weight and dilution of the sample) was vortex mixing of peritoneal fluid added into mass spectrometry water and then taken 100 μL to add into 300 μL of precipitant containing the mixed internal standard (acetonitrile: methanol=8:2), next vortexed and mixed it well, let it stand on the ice for 30 min, finally centrifuged it for 10 min at 4 °C at 12,000 rpm, and then take the supernatant of the whole sample for analysis by liquid chromatograph-mass spectrometer (LC-MS). Bile acids were quantified by ultra-performance liquid chromatography-tandem mass spectrometry (UHPLC-MS/MS) (ExionLC™ AD UHPLC-QTRAP 6500+, AB SCIEX Corp., Boston, MA, USA). The separation was performed on a Waters ACQUITY UPLC BEH C18 column (2.1 × 100 mm, 1.7 μm) with the maintained temperature at 50°C. The mass spectrometer was operated in negative multiple reaction mode (MRM). Raw data files generated by UHPLC-MS/MS were processed using Compound Discoverer 3.1 (CD3.1, Thermo Fisher) for peak alignment, peak pickup, and quantification of each metabolite. Subsequent visualization of the results was performed on R language (R4.3.2).
16s rRNA Sequencing Analysis
16S rRNA sequencing was employed to investigate whether there is dysbiosis in the gut microbiota of EM model mice. Samples were resuspended in Qiagen’s ASL buffer and homogenized for 2 min. Total fecal DNA was extracted from the supernatant using the ZR Fecal RNA kit (Zymo Research, USA). The concentration and purity of the extracted DNA were measured using NanoDrop 2000, and the quality of the DNA extraction was assessed by 1% agarose gel electrophoresis. Subsequently, polymerase chain reaction (PCR) amplification (forward primer: ACTCCTACGGGGAGGCAGCA; reverse primer: GGACTACHVGGGTWTCTAAT) was conducted by targeting the variable region V3-V4 of the 16S rRNA gene. Multiplex sequencing of amplicons with sample-specific barcodes was carried out using the Illumina NovaSeq platform. Quality control of the raw sequenced sequences was conducted using Trimmomatic software, and double-ended reads were then merged and spliced using FLASH v.1.2.7. The merged sequences were analyzed using QIIME v.1.9.1 software.30 The merged clean amplicons were clustered into operational taxonomic units (OTUs) using the “OpenReference” clustering method at a 97% similarity threshold. The clustered representative sequences were annotated with species using the RDP classifier and compared to Greengenes version 13.5.31 The obtained FEATURE TABLE and OTU annotation results were imported into R for subsequent visualization and analysis.
Single-Cell Analysis
Single-cell and spatial transcriptomic analysis were utilized to explore the targets of IL-17A/IL-17RA from both single-cell level and spatial ecological perspectives. The GSE179640 dataset from the GEO database was analyzed using the R package Seurat 4.0,32 imported as a Seurat object. Further processing of the resulting count matrices excluded genes detected in fewer than 3 cells and cells with (1) fewer than 500 genes, (2) greater than 10,000 genes, and (3) a maximum mitochondrial content of 25%. Two steps were skipped since the original dataset reported that the cell cycle and doublet had little effect on the results. Sample integration was then performed using the Harmony package to minimize batch effects. Seurat’s FindNeighbors function was then used with 15 dimensions as a parameter, and the FindClusters function was applied with a resolution of 3. The AddModuleScore function included in the Seurat package was used to assess the scores of signaling pathways in different cell subpopulations. Pathway analysis was conducted using the ClusterProfiler R package v.4.4.2.
Spatial Transcriptome Analysis
Raw data and H&E images of GSM6690475 from GEO Data were downloaded.25 Space Ranger (v.1.2.1) was used for comparison and quality control. Spatial transcriptome data was imported using the Seurat package. To identify the predominant cell type in each spot, here we used the RCTD33 (robust cell-type decomposition) approach to annotate the cell-type composition at the spatial locus. RCTD simply involves decomposing spatial cell-type mixtures from cell-type profiles learned from single-cell RNA-seq while correcting for differences between sequencing technologies. The proportion of cells on each spot is subsequently labeled at the spatial locus for subsequent analysis.
Statistical Analysis
The selection of sample sizes for human follicular fluid and peritoneal fluid was based on previous studies and determined by efficacy analysis using StatMate version 2.0 (GraphPad Software), without excluding any data. Statistical analyses were performed using Prism GraphPad software v.9.0. Error bars indicate the standard error of the mean for all plots, and p-values were determined by a two-tailed student test or ANOVA. Two-sided p-values <0.05 were considered statistically significant. In single-cell analysis, all hypothesis tests were performed using the Wilcoxon rank sum test unless otherwise indicated, and the Benjamini-Hochberg correction was used to correct for multiple simultaneous hypothesis tests when applicable.
Results
IL17RA and the IL-17 Signaling Pathway Act Primarily on Myeloid Cells in Peritoneal Lesions of EM
To investigate the potential role of the IL-17 and IL-17 signaling pathway in EM, we analyzed scRNA-seq datasets from 14 individuals obtained from a public database (GSE179640). These datasets encompassed four different locations, including endometrial samples from three healthy females (Ctrl) and samples from 11 EM patients, which were categorized into eutopic endometrium (EuE), ectopic peritoneal lesions (EcP), ectopic peritoneal adjacent areas (EcPA), and ectopic ovarian lesions (EcO, Figure S2A and S2B)). Based on previously reported marker genes, the main cell types were classified as epithelial cells (EPCAM, CDH1), stromal cells (COL1A1, PDGFRA), smooth muscle cells (ACTA2), endothelial cells (VWF, PECAM1), lymphoid cells (CD2, CD3G), and myeloid cells (CD68, CD14, Figure 1A and B).
To explore how the IL-17A/IL-17RA axis functions across various cell types, we utilized the “IL-17 signaling pathway” gene set to score cell types, revealing notably high scores in myeloid cells (Figure 1C). Subsequently, we analyzed the expression levels of the IL-17 family and its receptors. Interestingly, we found significant expression of IL17RA only in myeloid cells (Figures S2C and S3A), while genes encoding IL-17 family cytokines (IL-17A, IL-17B, IL-17C, and IL-17D) were predominantly unexpressed in the endometrium and EM lesions, suggesting that elevated IL-17A levels in EM may originate from elsewhere, and myeloid cell populations may represent the primary targets of IL-17. Further comparisons of IL17RA levels across different tissues and their correlation with different cell types revealed higher IL17RA levels in EuE, EcP, and EcPA compared to Ctrl, with a significant association between myeloid cell proportions and EcP and EcPA (Figure 1D and E). These findings suggest a significant enrichment of myeloid cells in EcP and imply a potential pivotal role of the IL17A/IL17RA axis in the progression of EcP.
IL17RA+ Myeloid Cells May Play a Role in Chemotactic Migration in Abdominal Lesions of EM
To further investigate the role of the IL-17A/IL-17RA axis in myeloid cells of EcP, myeloid cells from EcP and EcPA were extracted and further divided into monocytes (Mono), macrophages (Marco), dendritic cells (DC), and mast cells (Figures 2A, B and S3B). Among these subsets of myeloid cells, the highest levels of IL17RA were found in Mono, whereas IL17RA was largely not expressed in mast cells (Figures 2C and S3C).
IL-17 signaling pathway gene set scoring of myeloid cell subpopulations also revealed higher scores for Mono and Macro compared to DC and mast cells (Figure 2E), as well as different genes in the IL-17 signaling pathway gene set with different expression levels in different myeloid cell subpopulations (Figure 2D), suggesting that the IL-17A/IL-17RA axis plays different biological roles for different myeloid cells in disease progression of EM. To explore the effect of myeloid cells with high IL17RA expression, myeloid cells were classified into two clusters of IL17RA-expressing (IL17RA+) and non-IL17RA-expressing (IL17RA−) to perform differential gene analysis (Figures 2F and S3D). Subsequently, the genes significantly expressed in IL17RA+ were analyzed by GO enrichment analysis, finding besides “positive regulation of cytokine production” significantly enriched, the pathways in “secretory granule membrane”, “leukocyte migration”, and “vesicle lumens” also enriched (Figure 2G). Moreover, GSEA analysis also showed chemotactic migration of IL17RA+ myeloid cells (Figure 2H). These results suggest that myeloid cells with high expression of IL17RA in EcP perhaps contribute to the progression of EM through pro-migration, secretion of vesicles, and production of inflammatory cytokines.
The Spatial Ecological Distribution of IL17RA+ Myeloid Cells in EcP is Revealed by Spatially Resolved Transcriptomics
The potential role of IL17RA+ myeloid cells in EcP was identified in the above study, but the spatial ecological niche relation to stromal or epithelial cells is not yet known. Thus, we explored their relationship through the public spatial transcriptome database (GSM6690475) of EcP. The RCTD (robust cell-type decomposition) method was used to annotate the cell-type composition at spatial spots.33 As shown in Figure 3A, RCTD assisted in identifying the proportion of cell types on each spot, and there were 1–10 cells on each spatial spot. The areas where epithelial cells were concentrated were mainly lesion locations (Figure 3A and B), whereas the remaining areas were mainly stromal cells. Myeloid cells were evenly distributed throughout the lesion (Figure 3C and D), suggesting the infiltration of myeloid cells in EcP. Additionally, the subset of IL17RA-expressing cells was distributed throughout the lesion along with myeloid cells as expected (Figure 3E). The expression of RORC, a major transcription factor encoding IL17A, was distributed throughout the lesions (Figure 3F), which presumably indicates the progression of lesions promoted by producing IL17A to induce myeloid cells to converge as its periphery.
Gut Microbiota Disorder Promotes Lesion Progression of EM and May Be Associated with IL17A
Our study revealed that the IL17A gene was scarcely detected in scRNA-seq of endometriotic lesion tissues. However, elevated levels of the cytokine IL-17A were detected in the peritoneal fluid and follicular fluid of EM patients (Figure 4A). It is well known that the gut microbiota is closely related to the systemic metabolic profile, and alterations in the microbiota affect changes in metabolic levels throughout the body.34 Disordered microbiota in EM was used to intervene in normal mice, resulting in inflammation in the peritoneal fluid of mice in our team’s previous study.26 As a result, we focused our goal on the effects of gut microbiota in EM and the intestinal environment on peritoneal IL-17A.
Previous reports have shown that intestinal tissues (colon and small intestine) have a higher percentage of Th17 cells compared to spleen and lymphoid tissues.18 Accordingly, the proportion of Th17 cells in the lamina propria in mice with EM was first examined, and found that the proportion of Th17 cells was significantly higher in mice with EM compared to normal mice (Figure 4B). Combined with our team’s previous report,4 gut microbiota disorder exists in patients as well as in mice with EM, presumably leading to dysregulation of gut homeostasis and intestinal wall damage. Significant differences in the gut microbiota of mice with EM compared to the normal group and increased gut permeability were shown in the present experimental data (Figure 4C and D). Moreover, we observed the mild inflammatory state in the gut of mice with EM (Figure 4E), which is consistent with previous studies in which rhesus monkeys with EM were more likely to have intestinal inflammation than controls.35
To further explore whether gut microbiota disorder promotes the progression of EM, the FMT experiment was performed (Figure 5A). The results showed a significant improvement in pelvic adhesion score, largest lesion volume, and number of lesions after mice with EM receiving normal mouse feces (FMT-CON), whereas mice receiving EM feces (FMT-EM) did not (Figure 5B-D). In addition, the expression of intestinal tight junction-related protein Claudin-1 in FMT-CON was also higher than in FMT-EM (Figure 5E), and intestinal permeability was significantly improved in the FMT-CON group (Figure 5F). These results suggest that gut microbiota disorder in EM compromises the intestinal wall barrier and promotes the progression of EM.
In summary, gut microbiota disorder in EM compromises the intestinal wall barrier and promotes the progression of EM, and may be a key factor in regulating Th17 cells in the intestine and increasing IL-17A levels in peritoneal fluid.
Chenodeoxycholic Acid (CDCA) and Ursodeoxycholic Acid (UDCA) Regulate Gut Microbiota and Intestinal Th17/Treg Cells in the EM
Abnormal levels of bile acids in the intestinal metabolites in mice with EM have been reported in our previous study.4 Consistent with previous studies, we found that CDCA and UDCA, a primary and a secondary bile acid, were significantly increased in the feces of mice with EM (Figure 6A).
Subsequently, CDCA and UDCA were separately used to intervene in mice with EM, exploring the effects on gut microbiota. The results showed that the gut microbiota of the UDCA group converged to the EM group at the OTU level, whereas the CDCA group did not overlap with the EM group and clustered toward the CON group (Figure 6B-D), suggesting that both CDCA and UDCA have the potential to regulate the gut microbiota of EM, but CDCA significantly improves the gut microbiota of EM and enables it to converge toward CON group.
Besides, we further detected the ratio of Th17 and Treg cells in the intestinal lamina propria after bile acid intervention due to the balance of Th17/Treg cell ratio regulates the autoimmune status and the imbalance of this ratio may cause intestinal inflammation in previous reports. As expected, there was an imbalance of Th17/Treg cells in mice with EM (Figure 6E). Interestingly, both CDCA and UDCA reduced the Th17 ratio in the intestinal lamina propria of EM, while the difference was not significant for improving the Treg cell ratio (Figure 6E).
UDCA May Promote EM Progression by Elevating Th17 Cell Levels in Peritoneal Fluid While CDCA Inhibits EM Progression
We explored the interventional effects of the two bile acids in EM mice. Surprisingly, only CDCA had an improved effect on EM (the therapeutic positive control was the Dienogest (DG) group), while UDCA had no therapeutic effect (Figure 7A-C). To investigate its mechanism, we found that CDCA improved the proportion of Th17 cells in the peritoneal fluid of mice with EM, but UDCA significantly increased the proportion of Th17 cells (Figure 7D).
Subsequently, bile acid metabolic profiles were found to be significantly enriched in the peritoneal fluid of mice with EM compared with normal mice by targeted bile acid metabolomics (Figure 7E), which may be due to the damage to the intestinal wall barrier and the entry of intestinal contents into the peritoneal cavity. Correlation analysis showed that bound UDCA was significantly correlated with CA, β-MCA, and GCA (Figure 7F) and the concentration of all of these bile acids correlated with UDCA was significantly elevated in peritoneal fluid of EM (Figure 7G). Recent studies have shown that the microbiota with similar variability has an effect on metabolites in the peritoneal fluid of mouse,34 inferring gut microbiota disorder of EM may affect the levels of various metabolites in the peritoneal fluid, resulting in the alteration of the immune microenvironment in the peritoneal cavity, which may contribute to the progression of EM.
Discussion
In recent years, the close relationship between EM and the microbiota from the vagina to the gut has been extensively studied, and yet the complex mechanisms underlying it are multifaceted and have not been fully elucidated. Our study proposed a new view of the association between gut microbiota and EM: gut microbiota disorder of EM may interfere with bile acid metabolism in the intestine, disturb immune homeostasis in intestinal lamina propria, and promote the release of cytokine IL-17A which may bind to IL-17RA expressed by myeloid cell populations infiltrated by the lesion and exerts its chemotactic effect to promote lesion progression.
Previous studies by our team have found significant differences in the gut microbiota of both women with EM and animal models compared to controls, characterized by a reduced diversity of gut microbiota and an increased phylum Firmicutes/Bacteroidetes ratio in EM.4,5 Inflammatory cytokines in the peripheral blood of EM were correlated with gut microbiota, as well as abnormal levels of bile acid metabolism in gut metabolites. Some studies have also identified associations between intestinal microbial metabolites and EM. For instance, α-linolenic acid and α-lipoic acid have been found to ameliorate EM progression, while β-glucuronidase has been implicated in promoting EM progression.26,36,37 Furthermore, a close relationship has been found between bile acid metabolism and the gut microbiota, that is bile acids remodel the structure of gut microbiota, and in turn, the gut microbiota regulates bile acid metabolism levels, and it is now confirmed that several secondary bile acids are metabolized by the gut microbiota.38 Primary bile acids produced by the liver and secondary bile acids derived from the gut microbiota together regulate the immune environment of entire intestinal or systemic immune homeostasis.39 A study has shown that secondary bile acids 3-oxoLCA and isoalloLCA which derive from the gut microbiota act as modulators of T cells in mice.13 3-OxoLCA inhibits the differentiation of Th 17 cell by directly binding to the key transcription factor retinoic acid-related orphan receptor-γt, whereas isoalloLCA increases the differentiation of Treg cell by generating mitochondrial reactive oxygen species.13 In contrast, the biosynthetic enzymes that produce the two bile acids are regulated by specific gut microbiota.14 These studies suggest a correlation between gut microbiota, bile acids and immunity.
Based on the above findings, our study revealed that gut microbiota disorder in EM probably be linked to EM lesions through bile acid metabolism and IL-17A. First, we found elevated levels of CDCA and UDCA in the intestines of mice with EM, as well as an abnormal proportion of Th17 cells in the intestinal lamina propria. The increase in CDCA is presumably due to the self-protection of the organism that is regulating the gut microbiota, inhibiting cholesterol synthesis (which is the raw material for the synthesis of estrogens), and improving EM. Although both CDCA and UDCA improved the proportion of Th17 cells in the intestinal lamina propria, UDCA significantly increased the proportion of Th17 cells in the peritoneal fluid of mice with EM because UDCA is more hydrophilic than CDCA and can easily enter the peritoneal cavity. UDCA has induced an increase of Th17 cells by directly contacting with T cells.
In addition, it has been shown that CDCA, as a precursor for the synthesis of UDCA, forms the secondary bile acid UDCA in the presence of hydroxysteroid dehydrogenase produced by the gut microbiota,40 suggesting that the gut microbiota perhaps control the immune environment of EM by regulating the CDCA/UDCA ratio. Besides, our study revealed that abnormalities in bile acid metabolism were also present in the peritoneal fluid of mice with EM, which had not been reported previously. These abnormal bile acids may perturb the immune environment in the peritoneal cavity, which in turn establishes a link between EM lesions.
Both this and previous studies have confirmed that IL-17A is increased in peritoneal fluid and follicular fluid of patients with EM.20 In recent years, studies have revealed the dual role of IL-17A in protective immunity and destructive inflammation, where it plays a key protective role in immunity against fungal, bacterial, and many viral and parasitic pathogens but can also mediate destructive infection-associated immunopathology, leading to the development of autoimmune or other chronic inflammatory diseases.15 In turn, EM as a chronic inflammatory disease results from a combination of immune dysregulation factors with a quite complex immunologic basis of lesions.41 The role of IL-17A in EM has not yet been fully clarified. Previous studies have shown that IL-17A promotes macrophage polarization toward M2 in the peritoneal fluid, exerting its immunosuppressive effect and promoting the growth of lesions.42 We found significantly elevated levels of IL17RA expression in infiltrating myeloid cells in peritoneal lesions of EM by single-cell analysis, and IL-17RA is a key receptor that binds to IL-17A. Furthermore, we found that myeloid cells with high IL17RA expression were functionally enriched for chemotactic migration. Myeloid cells, particularly macrophages, have been identified as a central component of the endometriosis ecosystem and play a key role in the establishment of endometriosis.43 Past studies have shown that the absence of IL17RA in myeloid cells improves liver tumorigenesis44 and the IL-17A/IL-17RA axis plays a proatherogenic role by regulating myeloid cell recruitment in aortic45 which suggests a pathogenic role for the myeloid cell IL-17A/IL-17RA axis. Overall, the role of the IL-17A/IL-17RA axis in EM needs to be confirmed by further studies, and blocking IL-17RA may be a future strategy for the treatment of EM.
However, there are some limitations of this study that need to be discussed at the end. First, this study lacks direct evidence to support the binding of IL-17A to myeloid IL-17RA and the subsequent biological role it plays. Considering the many studies documenting the mechanism of EM, further precise studies are needed. Second, further studies are needed to determine whether bile acid metabolism levels are abnormal in the peritoneal fluid of patients with EM.
Conclusion
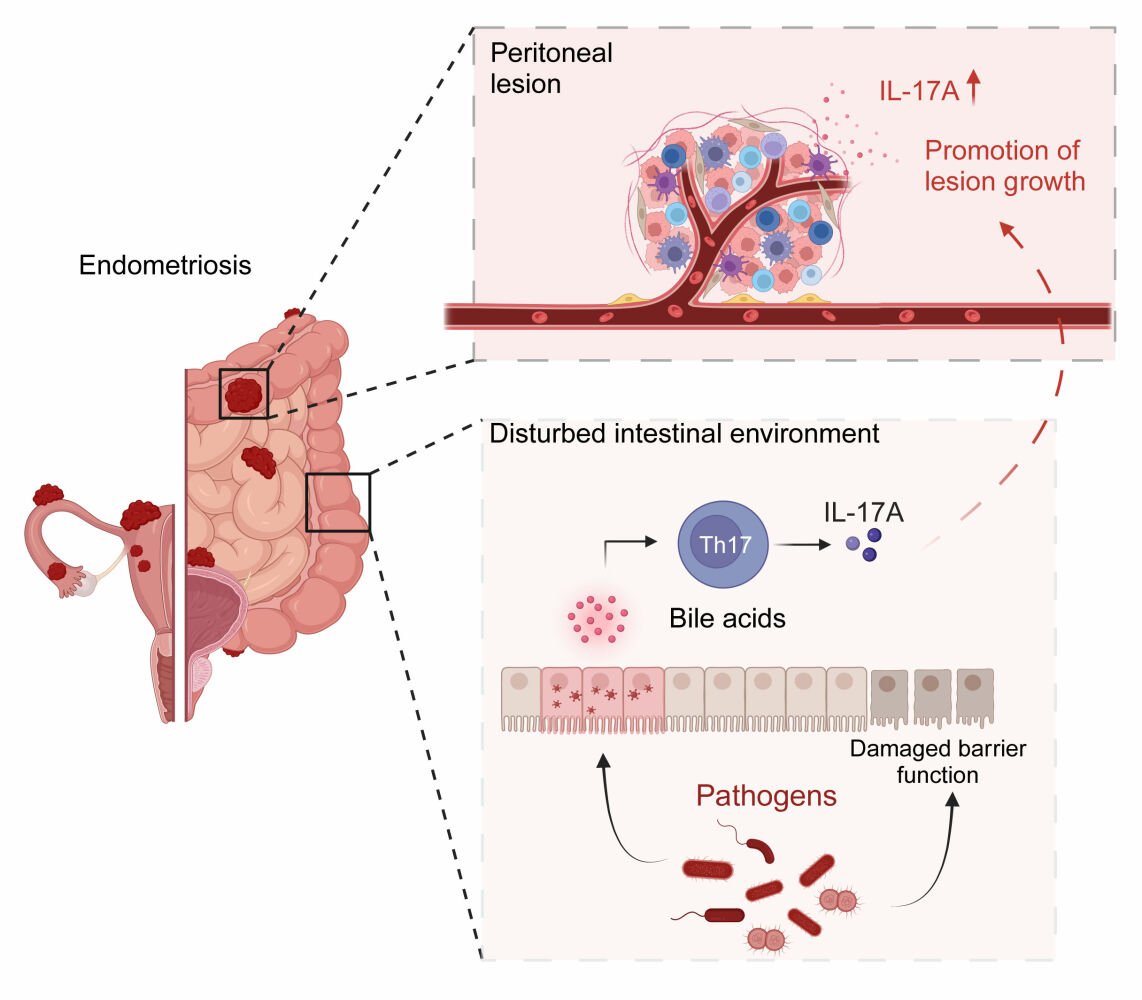
In conclusion, we present evidence that gut microbiota disorder in EM may impair the intestinal immune environment and bile acid metabolism. The IL-17A/IL-17RA axis may be an important factor promoting the progression of EM, and targeted blockade of it may be an option for the treatment of patients with EM in the future (Figure 8).
 |
Figure 8 Schematic illustrating the proposed pathogenic mechanism of disrupted gut microbiota in endometriosis Created with biorender.com. |
Acknowledgments
This work was supported by the National Natural Science Foundation of China (82374503, 82074206), Shanghai Municipal Science and Technology Commission “Science and Technology Innovation Action Plan” Medical Innovation Research Special Fund (21Y21920500) and First Affiliated Hospital of Naval Medical University “234 Discipline Climbing Program” (2020YXK007).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Zondervan KT, Becker CM, Missmer SA. Endometriosis. New Engl J Med. 2020;382(13):1244–1256.
2. Chapron C, Marcellin L, Borghese B, Santulli P. Rethinking mechanisms, diagnosis and management of endometriosis. Nat Rev Endocrinol. 2019;15(11):666–682.
3. Bulun SE. Endometriosis. New Engl J Med. 2009;360(3):268–279.
4. Ni Z, Sun S, Bi Y, et al. Correlation of fecal metabolomics and gut microbiota in mice with endometriosis. Am J Reprod Immunol. 2020;84(6):e13307.
5. Shan J, Ni Z, Cheng W, et al. Gut microbiota imbalance and its correlations with hormone and inflammatory factors in patients with stage 3/4 endometriosis. Arch Gynecol Obstet. 2021;304(5):1363–1373.
6. Tirone C, Pezza L, Paladini A, et al. Gut and Lung Microbiota in Preterm Infants: immunological Modulation and Implication in Neonatal Outcomes. Front Immunol. 2019;10:2910.
7. Wang L, Cai Y, Garssen J, Henricks PAJ, Folkerts G, Braber S. The Bidirectional Gut-Lung Axis in Chronic Obstructive Pulmonary Disease. Am J Respir Crit Care Med. 2023;207(9):1145–1160.
8. Cryan JF, O’Riordan KJ, Cowan CSM, et al. The Microbiota-Gut-Brain Axis. Physiol Rev. 2019;99(4):1877–2013.
9. Inversetti A, Zambella E, Guarano A, Dell’Avanzo M, Di Simone N. Endometrial Microbiota and Immune Tolerance in Pregnancy. Int J Mol Sci. 2023;24(3).
10. Jia W, Li Y, Cheung KCP, Zheng X. Bile acid signaling in the regulation of whole body metabolic and immunological homeostasis. Sci China Life Sci. 2024;67(5):865–878.
11. Bachem A, Makhlouf C, Binger KJ, et al. Microbiota-Derived Short-Chain Fatty Acids Promote the Memory Potential of Antigen-Activated CD8(+) T Cells. Immunity. 2019;51(2):285–297.e285.
12. Schneider KM, Blank N, Alvarez Y, et al. The enteric nervous system relays psychological stress to intestinal inflammation. Cell. 2023;186(13):2823–2838.e2820.
13. Hang S, Paik D, Yao L, et al. Bile acid metabolites control T(H)17 and T(reg) cell differentiation. Nature. 2019;576(7785):143–148.
14. Paik D, Yao L, Zhang Y, et al. Human gut bacteria produce Τ(Η)17-modulating bile acid metabolites. Nature. 2022;603(7903):907–912.
15. Mills KHG. IL-17 and IL-17-producing cells in protection versus pathology. Nat Rev Immunol. 2023;23(1):38–54.
16. Schnell A, Littman DR, Kuchroo VK. T(H)17 cell heterogeneity and its role in tissue inflammation. Nat Immunol. 2023;24(1):19–29.
17. Guo M, Liu H, Yu Y, et al. Lactobacillus rhamnosus GG ameliorates osteoporosis in ovariectomized rats by regulating the Th17/Treg balance and gut microbiota structure. Gut Microbes. 2023;15(1):2190304.
18. Schnell A, Huang L, Singer M, et al. Stem-like intestinal Th17 cells give rise to pathogenic effector T cells during autoimmunity. Cell. 2021;184(26):6281–6298.e6223.
19. Shi JL, Zheng ZM, Chen M, Shen HH, Li MQ, Shao J. IL-17: an important pathogenic factor in endometriosis. Int J Med Sci. 2022;19(4):769–778.
20. Ahn SH, Edwards AK, Singh SS, Young SL, Lessey BA, Tayade C. IL-17A Contributes to the Pathogenesis of Endometriosis by Triggering Proinflammatory Cytokines and Angiogenic Growth Factors. J Immunol. 2015;195(6):2591–2600.
21. Hirata T, Osuga Y, Hamasaki K, et al. Interleukin (IL)-17A stimulates IL-8 secretion, cyclooxygensase-2 expression, and cell proliferation of endometriotic stromal cells. Endocrinology. 2008;149(3):1260–1267.
22. Hirata T, Osuga Y, Takamura M, et al. Interleukin-17F increases the secretion of interleukin-8 and the expression of cyclooxygenase 2 in endometriosis. Fertil Sterility. 2011;96(1):113–117.
23. Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol. 2018;18(1):35–45.
24. Tan Y, Flynn WF, Sivajothi S, et al. Single-cell analysis of endometriosis reveals a coordinated transcriptional programme driving immunotolerance and angiogenesis across eutopic and ectopic tissues. Nat Cell Biol. 2022;24(8):1306–1318.
25. Fonseca MAS, Haro M, Wright KN, et al. Single-cell transcriptomic analysis of endometriosis. Nat Genet. 2023;55(2):255–267.
26. Ni Z, Ding J, Zhao Q, et al. Alpha-linolenic acid regulates the gut microbiota and the inflammatory environment in a mouse model of endometriosis. Am J Reprod Immunol. 2021;86(4):e13471.
27. Schoultz I, Å V K. The Intestinal Barrier and Current Techniques for the Assessment of Gut Permeability. Cells. 2020;9(8).
28. Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol. 2014;14(10):667–685.
29. Leonardi I, Gao IH, Lin WY, et al. Mucosal fungi promote gut barrier function and social behavior via Type 17 immunity. Cell. 2022;185(5):831–846.e814.
30. Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336.
31. McDonald D, Price MN, Goodrich J, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. Isme j. 2012;6(3):610–618.
32. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420.
33. Cable DM, Murray E, Zou LS, et al. Robust decomposition of cell type mixtures in spatial transcriptomics. Nat Biotechnol. 2022;40(4):517–526.
34. Brown K, Thomson CA, Wacker S, et al. Microbiota alters the metabolome in an age- and sex- dependent manner in mice. Nat Commun. 2023;14(1):1348.
35. Bailey MT, Coe CL. Endometriosis is associated with an altered profile of intestinal microflora in female rhesus monkeys. Hum Reprod. 2002;17(7):1704–1708.
36. Wei Y, Tan H, Yang R, et al. Gut dysbiosis-derived β-glucuronidase promotes the development of endometriosis. Fertil Sterility. 2023;120(3 Pt 2):682–694.
37. Di Nicuolo F, Castellani R, Cicco Nardone A D, et al. Alpha-Lipoic Acid Plays a Role in Endometriosis: new Evidence on Inflammasome-Mediated Interleukin Production, Cellular Adhesion and Invasion. Molecules. 2021;26(2).
38. Cai J, Sun L, Gonzalez FJ. Gut microbiota-derived bile acids in intestinal immunity, inflammation, and tumorigenesis. Cell Host Microbe. 2022;30(3):289–300.
39. Su X, Gao Y, Yang R. Gut microbiota derived bile acid metabolites maintain the homeostasis of gut and systemic immunity. Front Immunol. 2023;14:1127743.
40. Li Y, Wang K, Ding J, Sun S, Ni Z, Yu C. Influence of the gut microbiota on endometriosis: potential role of chenodeoxycholic acid and its derivatives. Front Pharmacol. 2022;13:954684.
41. Symons LK, Miller JE, Kay VR, et al. The Immunopathophysiology of Endometriosis. Trends Mol Med. 2018;24(9):748–762.
42. Miller JE, Ahn SH, Marks RM, et al. IL-17A Modulates Peritoneal Macrophage Recruitment and M2 Polarization in Endometriosis. Front Immunol. 2020;11:108.
43. Saunders PTK, Horne AW. Endometriosis: etiology, pathobiology, and therapeutic prospects. Cell. 2021;184(11):2807–2824.
44. Ma HY, Yamamoto G, Xu J, et al. IL-17 signaling in steatotic hepatocytes and macrophages promotes hepatocellular carcinoma in alcohol-related liver disease. J Hepatol. 2020;72(5):946–959.
45. Butcher MJ, Gjurich BN, Phillips T, Galkina EV. The IL-17A/IL-17RA axis plays a proatherogenic role via the regulation of aortic myeloid cell recruitment. Circ Res. 2012;110(5):675–687.
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.







